the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Calpain 1 gene expression in liver tissue and the association of novel calpain 1 single-nucleotide polymorphisms (SNPs) with meat quality in Bali cattle
Dairoh
Mokhamad Fakhrul Ulum
Jakaria
Cece Sumantri
Calpain 1 (CAPN1) is an enzyme that influences meat tenderization, and it is involved in post mortem proteolysis. The bovine CAPN1 gene was chosen as a candidate gene for DNA sequencing to identify novel single-nucleotide polymorphisms (SNPs) in exons 8–10 and assess their associations with meat quality in Bali cattle. In an analysis of 95 Bali cattle, two novel SNPs (g.5327C>T and g.5959C>T) were identified in exons and four novel SNPs (g.5534C>T, g.5807A>C, g.5857G>A, and g.5869T>C) were discovered in introns. Allelic frequency was evaluated, and Hardy–Weinberg equilibrium was found for all SNPs in Bali cattle. The marbling score and intramuscular fat content as determined by ultrasound were associated with g.5869T>C and g.5959C>T. g.5327C>T was associated with pH and meat color in Bali cattle, whereas two other SNPs (g.5869T>C and g.5959C>T) were associated with Warner–Bratzler shear force and meat color. Furthermore, g.5869T>C was associated with linolenic acid content, and g.5959C>T with caprylic and linolenic acid levels. However, intronic SNPs (g.5534C>T, g.5807A>C, and g.5857G>A) did not significantly affect meat quality in Bali cattle. Quantitative real-time PCR of liver tissue revealed that the mRNA expression of CAPN1 significantly differed (P<0.05) among the CT, CC, and TT genotypes. The results suggest that genetic variability in loci within CAPN1 might be associated with meat quality in Bali cattle.
- Article
(789 KB) - Full-text XML
- BibTeX
- EndNote
The market for high-yielding beef cattle breeds has been affected by the recent large growth in the demand for beef, which is a crucial source of protein (Smith et al., 2018). Bali cattle is native Indonesian domesticated cattle (Martojo, 2012) that is used to produce good-quality meat. Bali cattle exhibit an extremely close kinship with the banteng (Bibos banteng), but they are distantly related to Bos taurus and Bos indicus based on an analysis of the D-loop mtDNA gene (Jakaria et al., 2019). The meat from Bali cattle is prized for its delicious taste and desirable texture. Consequently, these cattle are favored both by local farmers and consumers who prioritize high-quality sustainable meat production. Bali cattle have a compact body conformation with a carcass presentation rate of up to 56 % (Hafid et al., 2019). Bali cattle can attain United States Department of Agriculture (USDA) quality standards for beef at a relatively young age, namely 1.5–2.5 years, highlighting their efficiency in meat production (Aditia et al., 2014). High production of class I back cuts, especially cube roll and strip loin, can be achieved using Bali cattle (Aditia et al., 2014).
The meat quality of Bali cattle has been improved through enhanced management practices. One such strategy is crossbreeding Bali cattle with other cattle, including Wagyu cattle, to obtain combinations of beneficial genes to increase livestock productivity and generate a new breed for long-term production. Although crosses between Wagyu and Angus have been achieved (Liu et al., 2021), no crossbreeding between Wagyu and Bali has been reported.
Meat quality has a significant impact on crucial aspects of consumer acceptance, including the level of satisfaction among consumers, producers, and the meat industry at large (Khasrad et al., 2017). Meat quality indicators that significantly influence consumer acceptance include flavor, tenderness, marbling, and color, all of which have an important economic value (Geletu et al., 2021). Therefore, improving the quality of Bali beef is essential for supporting food security in Indonesia and ensuring the availability of high-quality meat that can meet the needs of the population. According to Berry and Kearney (2018), environmental factors such as feed quality, age at slaughter, management practices, and processing at the time of slaughter affect meet quality and genetic factors also have a significant influence on meat quality. Many genes affect the quantitative characteristics of meat. Selection based on phenotypic data is considered less efficient because environmental influences are high. Therefore, the marker-assisted selection method exhibits excellent potential because of its time and cost efficiencies and high accuracy (Nayak and Singh, 2017). In marker-assisted selection, molecular markers are used to map and identify genetic loci associated with traits of interest, thereby enabling more precise and focused genetic selection. Screening potential parents based on genetic information can help minimize environmental influences, increase selection precision, and accelerate genetic progress (Goddard, 2009).
In recent years, several genetic markers associated with variations in beef quality have been identified. Genome-wide association studies have been conducted to identify putative genes for meat quality. The proteolytic calpain system consists of the genes calpain 1 (CAPN1) and calpastatin (CAST), both of which required a high calcium concentration to function. Meat quality traits, particularly tenderness, are associated with CAPN1 (Mateescu et al., 2017). Myofibril proteins are degraded by calpain through post mortem proteolysis (Coria et al., 2018). Certain CAST enzymes suppress the proteolysis of calpain, making it inactive when calcium is present. Based on research conducted by Dairoh et al. (2021), the CAPN1 gene has been consistently demonstrated to affect the meat quality of Bali cattle. In their study, six single-nucleotide polymorphisms (SNPs) and eight indels in the CAPN1 gene were found to significantly affect variables related to meat quality, and these genetic variations were not found in B. indicus or B. taurus (Dairoh et al., 2022).
This study investigated the effects of SNPs within the CAPN1 gene on meat quality in Bali cattle. Through an analysis of SNP genetic diversity and an examination of the associations of SNPs with meat quality traits, specific genotypes influencing these aspects were identified. The second aim was to determine the association of CAPN1 gene expression with tenderness and fatty acid composition in Bali cattle. The findings of this research hold significant implications for the advancement and selection of Bali cattle. These findings will contribute to progress in genetic breeding practices aimed at enhancing meat quality within the Bali cattle population.
2.1 Blood sampling
Blood samples from 95 Bali cattle were collected in Kupang, East Nusa Tenggara province, Indonesia, and shipped to a Basirih slaughterhouse. The cattle were slaughtered at the Basirih slaughterhouse in south Banjarmasin, South Kalimantan province. Venoject was used to collect blood samples from the jugular vein of each animal, and samples were stored in a 1.5 mL EDTA vacuum container. This experimental procedure was approved by the Animal Ethics Committee of the Banjarmasin City Food Security, Agriculture, and Fisheries Service (approval no. 520/624/DKP3/X11/2021). DNA extraction was performed at the Animal Molecular Genetics Laboratory, Faculty of Animal Husbandry, IPB University. The DNA extraction method using the Geneaid kit was used for extraction.
2.2 Marbling score and intramuscular fat measurement
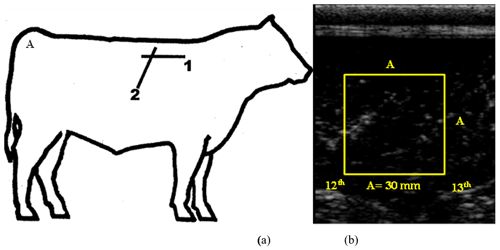
A 7.1 MHz ultrasound transducer was used to measure the marbling score and intramuscular fat content at the 12th–13th rib position (Ulum et al., 2014; BIF, 2016). Sonograms of intramuscular fat were obtained 3 d before slaughter. To prepare for scanning, the skin of living Bali cattle was shaved. Ultrasound gel was used to allow ultrasound waves to pass from the probe to the tissue of the cattle. The images were immediately processed on the monitor screen, and a picture of the transversal intramuscular fat content was displayed. Ultrasound images were saved and analyzed using ImageJ software (NIH, Bethesda, MD, USA; Fig. 1). The Australian Meat Standard was used to determine the marbling score.
2.3 Physical analysis
In total, 250 g of meat was taken from the tenderloin of 44 Bali cattle. The assessed physical characteristics included pH, tenderness, cooking loss, color, and water-holding capacity (WHC). pH was analyzed using a calibrated pH meter. For tenderness and cooking loss analysis, 100 g of meat was boiled until the temperature reached 80 °C. Meat samples were treated with a 1.27 cm beef corer to make small cuts and had their Warner–Bratzler shear force (WBSF) measured. The difference in weight between fresh and cooked meat was used to calculate cooking loss. The meat color was analyzed using red litmus paper. The press method was used to determine WHC. For the analysis, Whatman filter paper was used. The wet area was the water area formed by the difference between the outer and inner circles absorbed by the paper because of the clamping method.
2.4 Fatty acid analysis
Fatty acid composition was determined using the Association of Official Analytical Chemists method (AOAC 2012). A chloroform–methanol solution was used to extract lipids from 40 g of meat. The recovered lipids were then transesterified to yield fatty acid methyl esters (FAMEs). The FAMEs were extracted with hexane solution, centrifuged, and dried. The resultant FAME products were dissolved in chloroform solution before being filtered to eliminate undesirable chemicals using solid-phase extraction. One microliter of FAMEs was placed in gas chromatography (GC) equipment. The isolated fatty acids were identified and quantified by mass spectrometry. The retention time and peak were recorded for each fatty acid component. Comparing the retention time to standards yields information on each fatty acid component. GC was used to identify saturated, monounsaturated, and polyunsaturated fatty acids.
2.5 Primer design and DNA amplification
CAPN1 primer sequences were designed from exon 8 to exon 10 using the GenBank sequence of the National Center for Biotechnology Information (NCBI) (access code AH009246.3). The length of the primer sequences was determined using the Primer3 and Primer-BLAST websites, a Multiple Primary Analyzer, and Primary Stats software. The primer sequences were as follows: forward 5′-GTG AAC TAC CAG GGC CAG AT-3′ and reverse 5′-TCG TAC AGG GTG GTG TTC CA-3′. The PCR product was 763 bp in length. The CAPN1 gene was amplified using an AB System thermocycler and the following PCR conditions: predenaturation at 95 °C for 1 min; 35 cycles of denaturation at 95 °C for 15 s, annealing at 57 °C for 15 s, extension at 72 °C for 10 s; and final extension at 72 °C for 3 min. PCR products were separated by 1 % agarose gel in electrophoresis and examined using a UV Transilluminator (Bio-Rad, Hercules, CA, USA).
2.6 DNA sequencing and genotyping
The amplification product was sequenced on an ABI PRISM with the BigDye Terminator kit v3.1 by 1st Base Laboratory Services (Selangor, Malaysia). FinchTV, BioEdit, and Molecular Evolutionary Genetic Analysis (MEGA X) were used to analyze the sequencing results. The SNP positions were named after the complete NCBI reference sequence calculated from the first base of the CAPN1 gene.
2.7 Extraction of total RNA and cDNA quantification
Total RNA was extracted from liver tissue using the RNeasy Fibrous Tissue Mini Kit based on the association (SNP g.5959C>T) with different meat tenderness and fatty acid composition phenotypes of Bali cattle. RNA was extracted from liver tissue as described by Khansefid et al. (2018). The liver is a central organ responsible for glycogen production and fatty acid metabolism. Meat tenderness is influenced by the inherent qualities of the meat, particularly the glycogen content. The liver is the primary organ for glucose production, contributing 85 %–90 % of the body's glucose supply and storing glycogen to regulate blood glucose levels (Bergman et al., 1974). The regulation of muscle glycogen levels affects the meat's ultimate pH, color, and tenderness through post mortem glycolytic changes (Rosenvold et al., 2001). Furthermore, fatty acid metabolism and synthesis are predominantly orchestrated within the liver, which is pivotal in lipid metabolism. This organ is responsible for synthesizing, catabolizing, and modifying fatty acids (Alves-Bezerra and Cohen, 2017). Various enzymes and proteins involved in fatty acid metabolism are synthesized in the liver, making it a crucial organ for studying the genetic regulation of these processes (Vallim and Salter, 2010). Therefore, it was reasonable to assess gene expression utilizing liver tissue when investigating tenderness and fatty acid composition phenotypes along with their associated processes.
Table 1Primer information for the bovine CAPN1 gene and a housekeeping gene, β-actin.

CAPN1: calpain 1; F: forward; R: reverse.
Three groups based on the genotypes at the SNP g.5959C>T (CC, CT, and TT) were created to compare the relationships of CAPN1 gene expression with tenderness and fatty acid levels. The t test was performed using SAS statistical software (SAS Institute, Cary, NC, USA) to measure the significance of differences among the three genotypes. Total RNA was treated using RNase-Free DNase (Promega, Madison, WI, USA), which was followed by RNA measurement using a spectrophotometer. The Transcriptor cDNA Synthesis Kit was used to convert RNA into cDNA. Using real-time PCR, cDNA was used to calculate CAPN1 gene expression. Real-time PCR was performed with the SYBR Green master kit and a reaction mixture containing 5 µL of the master kit, 0.5 µL of the forward and reverse primers each (Table 1), 1 µL of sample cDNA, and 3 µL of NFW. Each cDNA sample and nontemplate control were examined in a 96-well microtiter plate in each run. The qRT-PCR machine was programmed to perform 5 min of predenaturation at 95 °C, 10 s of denaturation at 95 °C, 20 s of annealing at 54 °C, 30 s of extension at 72 °C, and 5 min of final extension at 72 °C. All samples were run in duplicate, and the target gene expression was normalized using the geometric mean of the housekeeping gene, β-actin. cDNA was quantified using the AG qTower Analytic Jena program (Listyarini et al., 2018).
Figure 2Construction of a fluorescence standard curve to calculate cycle threshold values by qRT-PCR.
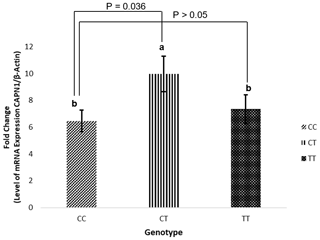
Figure 3CAPN1 mRNA expression in liver tissue based on genotype (CC, CT, and TT). mRNA expression is presented as the fold change using the β-actin gene as a reference. (CAPN1: calpain 1.)
CAPN1 gene expression as determined by qRT-PCR was based on the absolute quantification method. This method utilizes a standard fluorescence curve to represent the number of cycles required for fluorescence to surpass the cycle threshold (Ct). The recorded Ct during the exponential amplification phase was used to calculate CAPN1 gene expression (Fig. 2). This process involves comparing the sample CT to the standard curve, enabling the determination of the relative concentration of the CAPN1 gene in the tested cDNA sample. Thus, the absolute quantification method allows for the direct and accurate measurement of gene expression in analyzed samples (Gamal and Ibrahim, 2024). The absolute quantification of gene expression by qRT-PCR was performed using ΔCt expressed as a fold change, and the housekeeping gene β-actin was used as the reference gene. ΔCt was calculated by subtracting the Ct of the target gene from that of the reference gene (Silver et al., 2006) as follows:
2.8 Data analysis
CAPN1 gene diversity was investigated using the POPGENE 1.32 program, allele frequencies, genotype frequencies, observed and expected heterozygosity values, and the Hardy–Weinberg balance approach. The general linear model (GLM) method was used to investigate the relationships of CAPN1 genotypes with the marbling score, intramuscular fat content, physical traits, and fatty acid levels. The mathematical formula for the GLM model is as follows:
where Yij is the phenotypic observation, μ is the overall mean, Gi is the genotype effect, and eij is the random error.
3.1 Newly discovered SNPs in the CAPN1 gene
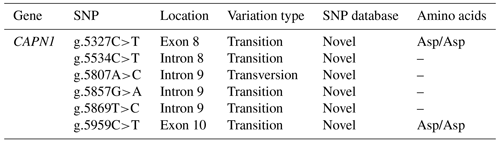
Six SNPs were identified in the CAPN1 gene using the chromatograms of double peaks from sequencing data. Two SNPs (g.5327C>T and g.5959C>T) were found in exons 8 and 10, respectively, whereas four SNPs (g.5534C>T, g.5807A>C, g.5857G>A, and g.5869T>C) were found in introns 8–9, as indicated in Table 2. Five SNPs (g.5327C>T, g.5534C>T, g.5857G>A, g.5869T>C, and g.5959C>T) in the CAPN1 gene were transition mutations and one (g.5807A>C) was a transversion mutation. These SNPs were entirely synonymous, causing no amino acid changes. However, no studies have explored the influence of these polymorphisms on phenotypic traits. Direct DNA sequencing identified the six CAPN1 SNPs as novel variations located between exons 8 and 10. The variants were was identified in the Bali sequence by aligning them with the entire ensemble (ENSBTAG00000010230) sequence of the cattle CAPN1 gene using MEGA X software.
Table 3Allelic and genotypic frequencies and diversity parameters of SNPs in the CAPN1 gene.
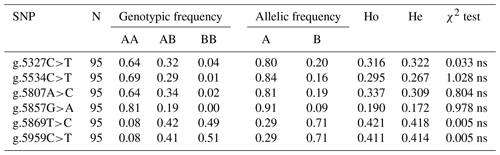
CAPN1: calpain 1; SNP: single-nucleotide polymorphism; AA: reference genotype (wild-type); AB: heterozygous genotype; BB: mutant genotype; Ho: observed heterozygosity; He: expected heterozygosity; ns: not significant (χ2 test) < chi-squared table (3.84: α0.05 df's 2).
3.2 CAPN1 gene polymorphism
Table 3 presents the allelic frequencies, genotype frequencies, heterozygosity (expected heterozygosity, He, and observed heterozygosity, Ho), and Hardy–Weinberg equilibrium of the CAPN1 gene. All SNPs in the CAPN1 gene had three genotypes: AA, AB, and BB. All six discovered SNPs were polymorphic, with minor allele frequencies exceeding 5 % (Allendorf et al., 2013). According to the genetic diversity analysis, the AA genotype was the most common for g.5327C>T, g.5534C>T, g.5807A>C, and g.5857G>A, whereas the BB genotype was the most common for g.5869T>C and g.5959C>T. Heterozygosity can be used to estimate a population's genetic diversity. Ho was higher than He for g.5869T>C and g.5959C>T. This suggests the existence of high genetic heterogeneity at specific loci (Fang et al., 2012). All SNPs in the CAPN1 gene were discovered to be in Hardy–Weinberg equilibrium. The Hardy–Weinberg equilibrium test is used to determine whether genotype proportions within a population remain constant throughout generations (Waples, 2015). The Bali cattle utilized in this investigation were obtained from breeders who used no selection and who did not deviate from the Hardy–Weinberg equilibrium (Garnier-Gere and Chikhi, 2013; Graffelman et al., 2017).
Table 4Association of CAPN1 gene SNPs with the marbling score and intramuscular fat percentage in Bali cattle.
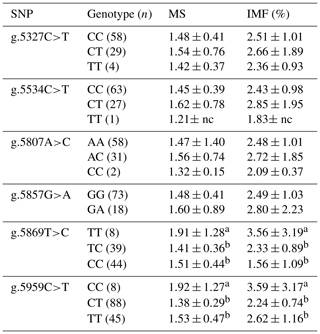
CAPN1: calpain 1; MS: marbling score; IMF: intramuscular fat; nc: not counted. a,b Means in the same column with different superscripts were significantly different (P<0.05). Values with the superscript a indicate different and higher values compared to those with the superscript b.
3.3 Association of CAPN1 gene SNPs with marbling and intramuscular fat
Table 4 displays the results of the associations of SNPs in the CAPN1 gene with the intramuscular fat percentage and marbling score of Bali cattle. The phenotypes of intramuscular fat and marbling were determined using ultrasonic scanning in Bali cattle. Two SNPs (g.5869T>C and g.5959C>T) in the CAPN1 gene were significantly associated (P<0.05) with the intramuscular fat percentage and marbling score. Marbling has been demonstrated to affect beef flavor, juiciness, tenderness, and visual qualities (Hwang et al., 2014; Silva and Cadavez, 2012). It also has a high heritability of 0.56. According to Park et al. (2018), a significant positive correlation (0.91) between tenderness and marbling supports the idea that marbling-based selection can significantly increase beef tenderness. Marbling is deposited within the endomysium, resulting in the weakening of the perimysial connective tissue and a more tender product (Nishimura, 2010). Prior studies demonstrated that several SNPs in the CAPN1 gene were significantly associated (P<0.05) with intramuscular fat content and marbling scores in Hanwoo and Nellore cattle (Cheong et al., 2008; Li et al., 2013). Previous research found that the SNP g.232G>C in the 5′ UTR of the CAPN1 gene is significantly associated (P<0.05) with the marbling score of Bali cattle (Dairoh et al., 2021). The findings confirm that genetic variants in CAPN1 serve as possible markers that can be used in breeding programs to selectively improve marbling scores and intramuscular fat content in beef cattle, positively contributing to beef quality.
Table 5Association of CAPN1 gene SNPs with physical traits in Bali cattle.
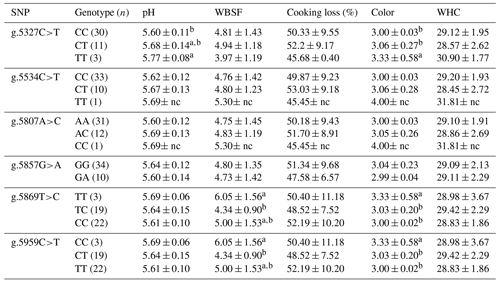
CAPN1: calpain 1; WBSF: Warner–Bratzler shear force; WHC: water-holding capacity; nc: not counted. a,b Means in the same column with different superscripts were significantly different (P<0.05). Values with the superscript a indicate different and higher values compared to those with the superscript b.
3.4 Association of CAPN1 gene SNPs with physical traits
The associations of CAPN1 gene polymorphisms with physical traits were analyzed in Bali cattle (n=44). The evaluated traits were pH, WBSF, cooking loss, meat color, and WHC. The results are presented in Table 5. g.5327C>T in the CAPN1 gene was significantly associated (P<0.05) with pH and meat color in Bali cattle. The association study revealed that g.5869T>C and g.5959C>T were significantly associated (P<0.05) with WBSF and meat color. Regarding the SNP g.5959C>T of the CAPN1 gene, Bali cattle with the CT genotype had significantly lower WBSF than those with the TT and CC genotypes (P<0.05). Tenderness was measured by shear force, with lower shear force values indicating greater tenderness. According to these findings, Bali cattle with the CT genotype of g.5959C>T were classified as tender, whereas those with the TT and CC genotypes were classified as harsh. Thus, the CT genotype is more desirable for high-quality meat. Tenderness is an essential meat quality attribute because variations in tenderness are suggested to influence consumers' decision to repurchase meat (Maltin et al., 2003). Several studies investigated the effect of genetic markers in the CAPN1 gene on beef tenderness (Tizioto et al., 2013; Magalhães et al., 2019; Bedhane et al., 2019). Previous studies revealed that SNPs in the CAPN1 gene are associated with tenderness (Curi et al., 2010; Sun et al., 2018). Therefore, these genetic markers might help select populations for beef breeding.
Prior studies demonstrated that the activity of proteolytic enzymes, including CAPN1 (also known as μ-calpain), influenced tenderness. Calpain can degrade muscle proteins such as titin, nebulin, vinculin, and desmin, which play roles in meat tenderness (Bhat et al., 2018). Calpain has greater ability to degrade the titin protein, which has a molecular weight (MW) of 2400 kDa, than it does to degrade titin, which has a MW of 3700 kDa (Iwanowska et al., 2011). In addition, the tenderness of meat can be influenced by pH and WHC during post mortem examination. Strong evidence has revealed that low pH post mortem was associated with pale meat and poor WHC, resulting in poor meat quality and particularly low tenderness. Rapid pH decline can cause proteolytic denaturation or autolysis, reducing tenderness (Santos et al., 2016).
Table 6Association of CAPN1 gene SNPs with the fatty acid composition in Bali cattle.
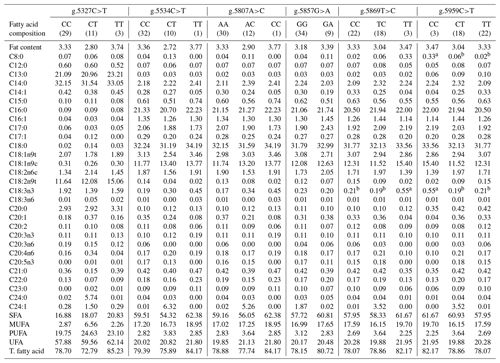
CAPN1: calpain 1; SFA: saturated fatty acid; MUFA: monounsaturated fatty acid; UFA: unsaturated fatty acid; T. fatty acid: total fatty acid; nc: not counted. a,b Means in the same row with different superscripts were significantly different (P<0.05). Values with the superscript a indicate different and higher values compared to those with the superscript b.
3.5 Association of CAPN1 gene SNPs with fatty acid content in Bali cattle
The associations of CAPN1 gene SNPs with the fatty acid composition of Bali cattle were examined, as presented in Table 6. Linolenic acid (C18:3n3) content was significantly associated with the intronic SNP, g.5869T>C (P<0.05). Specifically, the linolenic acid (C18:3n3) content was substantially greater in Bali cattle with the CC genotype than in cattle with the CT and TT genotypes. Linolenic acid (C18:3n3) is an omega-3 fatty acid and an essential fat that can only be obtained in food. Omega-3 fatty acids serve as substrates for the production of longer-chain omega-3 fatty acids, such as docosahexaenoic acid (C22:6 n-3) and eicosapentaenoic acid (C20:5 n-3), which are crucial for controlling blood pressure, inflammation, immune responses, brain development, and cognitive function (Sirot et al., 2008; Whitmore et al., 2019). Furthermore, omega-3 fatty acids have been demonstrated to contribute to preventing cardiovascular disease, diabetes, hypertension, allergies, cancer, renal disorders, and neural dysfunction and improving immune responses (Sinclair, 2019; Sokoła-Wysoczańska et al., 2018; Djuricic and Calder, 2021). Previous research found that SNPs in the exon 7 region of the CAPN1 gene were associated with myristic, palmitic, linoleic, and linolenic acid levels in Yanbian cattle (Xin et al., 2011). A previous study also reported that several polymorphisms in the SCARB1 and SREBF1 genes were significantly associated (P<0.05) with omega-3 fatty acid levels in Bali cattle (Dairoh et al., 2023).
Furthermore, the current study observed significant associations (P<0.05) of the exon 10 SNP, g.5959C>T, with caprylic acid (C8:0) and linolenic acid (C18:3n3) levels. Caprylic acid (C8:0), a medium-chain fatty acid, has been proven to reduce body weight while improving internal fat accumulation and cholesterol metabolism (Rego-Costa et al., 2012). Previously, Roopashree et al. (2021) indicated that medium-chain saturated fatty acids are utilized to treat several conditions, including high weight, high cholesterol levels, cardiovascular diseases, and Alzheimer's disease, and used for their immunomodulatory effects. According to the present study, beef cattle with desired SNPs in the CAPN1 gene might have high levels of fatty acids linked to selection. Furthermore, the newly discovered SNPs in these genes can be a great source of knowledge for enhancing meat quality, especially regarding fatty acids. More research in larger populations and diverse settings is necessary to validate the impact of the identified genetic variants.
3.6 Total mRNA expression of the CAPN1 gene in Bali cattle
CAPN1 gene expression in liver tissue was classified according to the CAPN1 g.5959C>T genotype. Tables 5 and 6 present the associations of g.5959C>T with WBSF and fatty acid content. The statistical results in Table 7 indicate that animals with the CT genotype had a higher mRNA expression than those with the CC genotype (P=0.036), whereas no difference was observed between the CT and TT genotypes or between the CC and TT genotypes. The increase in CAPN1 gene expression for the CT genotype indicated that muscle degradation was accelerated in Bali cattle, resulting in a lower shear force value (greater tenderness). Coria et al. (2020) found that greater shear force was associated with reduced CAPN1 gene expression. Previous research revealed that the higher shear force of zebu meat was most likely attributable to enhanced CAST expression rather than decreased expression of protease-encoding genes (Giusti et al., 2013). The present study also found that the CT genotype was associated with greater mRNA expression and decreased accumulation of linolenic acid (C18:3n3) and caprylic acid (C8:0). These findings indicate that the CAPN1 gene might influence meat quality and fatty acid content in Bali cattle. This finding could be used in future studies to identify the genetic basis for improved tenderization. Furthermore, the degree of CAPN1 expression under shear force needs to be clarified in future research on diverse tissues and a greater number of Bali cattle.
In this study, six new SNPs in the CAPN1 gene were found to be polymorphic in Bali cattle. SNPs g.5869T>C and g.5959C>T were significantly associated with the intramuscular fat percentage, marbling score, WBSF, meat color, and caprylic acid (C8:0) and linolenic acid (C18:3n3) content. Some beneficial features were observed in animals with the CT genotype of the SNP g.5959C>T. The CT genotype was linked to lower shear force (greater tenderness) and lower caprylic acid content (C8:0). The findings for the CT genotype differed significantly from those for the CC and TT genotypes. CAPN1 gene polymorphisms could be used as markers for the intramuscular fat percentage, marbling score, tenderness, and fatty acid composition in Bali cattle.
The datasets used in this study are available upon request from the author.
D was in charge of the experiment, data analysis, and writing the first draft. J, MFU, and CS were in charge of study design, data resources, and data validation. The submitted paper was read and approved by all authors.
The contact author has declared that none of the authors has any competing interests.
This experiment had no negative consequences for animal welfare. All procedures in this study were approved by the Banjarmasin City Food Security, Agriculture, and Fisheries Agency's Animal Ethics Committee (approval ID: 520/624/DKP3/XII/2021).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research was financially supported by Postdoctoral Project 2024 of IPB University (contract no. 9/IT3.D12/SP/DAPT.PD/2024), and National Collaborative Research (contract no. 504/IT3.D10/PT.01.03/P/B/2023).
This research has been supported by the Postdoctoral Project of IPB University (contract no. 9/IT3.D12/SP/DAPT.PD/2024) and the National Collaborative Research RI-NA project (contract no. 504/IT3.D10/PT.01.03/P/B/2023).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Aditia, E. L., Priyanto, R., Baihaqi, M., Putra, B. W., and Ismail, M.: Carcass characteristics of bali and ongole crossbreed cattle fed with sorghum base, Proceedings of The 16th AAAP Animal Science Congress Vol. II, Sustainable Livestock Production in the Perspective of Food Security, Policy, Genetic Resources and Climate Change, Yogyakarta, 10–14 Nopember 2014, Yogyakarta, Indonesia, 911–914, ISBN 978-602-8475-87-7, 2014.
Allendorf, F. W., Luikart, G. H., and Aitken, S. N.: Conservation and the genetics of population, Edisi ke-2, Willey-Blackwell, New York, United States, ISBN 978-0-470-67145-0, 2013.
Alves-Bezerra, M. and Cohen, D. E.: Triglyceride metabolism in the liver, Compr. Physiol., 8, 1–8, https://doi.org/10.1002/cphy.c170012, 2017.
Bedhane, M., Werf, J., Gondro, C., Duijvesteijn, N., Lim, D., Park, B., Hee, R. S., and Clarck, S.: Genome-wide association study of meat quality traits in Hanwoo beef cattle using imputed whole-genome sequence data, Frontiers in Genetics, 10, 1235, https://doi.org/10.3389/fgene.2019.01235, 2019.
Bergman, E. N., Brockman, R. P., and Kaufman, C. F.: Glucosa metabolism in ruminants: comparison of whole-body turnover with production by gut, liver, and kidneys, Fed. Proc., 33, 1849–1854, 1974.
Berry, D. P. and Kearney, J. F.: Development and implementation of national breeding programs for dairy and beef cattle in Ireland, J. Anim. Sci., 96, 3757–3764, 2018.
Bhat, Z. F., Morton, J. D., Mason, S. L., and Bekhit, A. E.: Role of calpain system in meat tenderness: a review, food science and human wellness, Food Sci. Hum. Wellness, 7, 196–204, https://doi.org/10.1016/j.fshw.2018.08.002, 2018.
Beef Improvement Program (BIF): Guidelines for uniform beef improvement program 9th Ed, North Carolina, Nort Carolina State University, P, 35–46, http://guidelines.beefimprovement.org/index.php?title=Guidelines_for_Uniform_Beef_Improvement_Programs&oldid=2716 (last access: 2 July 2024), 2016.
Cheong, H. S., Yoon, D. H., Park, B. L., Kim, L. H., Bae, J.S., Namgoong, S., Lee, H. W., Han, C. S., Kim, J. O., Cheong, I. C., and Shin, H. D.: A single nucleotide polymorphism in CAPN1 associated with marbling score in Korean cattle, BMC Genet., 9, 33–39, https://doi.org/10.1186/1471-2156-9-33, 2008.
Coria, M. S., Pedro, G. C., and Gustavo, A. P.: Calpain System in meat tenderization: a molecular approach, Rev. MVZ Cordoba, 23, 6523–6536, 2018.
Coria, M. S., Reineri, P. S., Pighin, D., Barrionuevo, M. G., Carranza, P. G., Grigioni, G., and Palma, G. A.: Feeding strategies alter gene expression of the calpain system and meat quality in the longissimus muscle of Braford steers, Asian Austral. J. Anim., 33, 753–762, https://doi.org/10.5713/ajas.19.0163, 2020.
Curi, R. A., Chardulo, L. A., Giusti, J., Silveira, A. C., Martins, C. L., and de Oliveira, H. N.: Assessment of GH1, CAPN1 and CAST polymorphisms as markers of carcass and meat traits in Bos indicus and Bos taurus-Bos indicus cross beef cattle, Meat Sci., 86, 915–920, https://doi.org/10.1016/j.meatsci.2010.07.016, 2010.
Dairoh, D., Jakaria, Ulum, M. F., Ishak, A. B. L., and Sumantri, C.: Association of SNP g.232 G>T calpain gene with growth and live meat quality prediction using ultrasound images in Bali cattle, Jurnal Ilmu Ternak dan Veteriner, 26, 49–56, https://repository.pertanian.go.id/handle/123456789/13342, 2021.
Dairoh, D., Jakaria, J., Ulum, M. F., and Sumantri, C.: A new SNP at 3'UTR region of calpain 1 gene and its association with growth and meat quality in beef cattle, Journal of the Indonesian Tropical Animal Agriculture, 47, 273–284, https://doi.org/10.14710/jitaa.47.1.17-28, 2022.
Dairoh, D., Ulum, M. F., Jakaria, Ishak, A. B. L., and Sumantri, C.: A novel snps of the srebf1 and scarb1 genes and the association with fatty acid profile in bali cattle, Tropical Animal Science Journal, 46, 428–438, https://doi.org/10.5398/tasj.2023.46.4.428, 2023.
Djuricic, I. and Calder, P. C.: Beneficial outcomes of omega-6 and omega-3 polyunsaturated fatty acids on human health, Nutrients, 13, 2421, https://doi.org/10.3390/nu13072421, 2021.
Fang, X., Zhang, J., Xu, H., Zhang, C., Du, Y., Shi, X., Chen, D., Sun, J., Jin, Q., Lan, X., and Chen, H.: Polymorphisms of diacylglycerol acyltransferase 2, gene and their relationship with growth traits in goats, Mol. Biol. Rep., 39, 1801–1807, https://doi.org/10.1007/s11033-011-0921-0, 2012.
Gamal, M. and Ibrahim, M. A.: Introducing the f0 % method: a reliable and accurate approach for qPCR analysis, BMC Bioinformatics, 25, 1–18, https://doi.org/10.1186/s12859-024-05630-y, 2024.
Garnier-Géré, P. H. and Chikhi, L.: Population subdivision, Hardy–Weinberg equilibrium and the Wahlund effect, New York, American Cancer Society, https://doi.org/10.1002/9780470015902.a0005446.pub3, 2013.
Geletu, U. S., Usmael, M. A., Mummed, Y. Y., and Ibrahim, A. M.: Quality of cattle meat and its compositional constituents, Veterinary Meidcine International, 2012, 1–9, https://doi.org/10.1155/2021/7340495, 2021.
Giusti, J., Castan, E., Dal Pai, M., Arrigoni, M. D. B., Rodrigues, B. S., and De Oliveira, H. N.: Expression of genes related to quality of longissimus dorsi muscle meat in Nellore (Bos indicus) and Canchim (5/8 Bos taurus × 3/8 Bos indicus) cattle, Meat Sci., 94, 247–252, https://doi.org/10.1016/j.meatsci.2013.02.006, 2013.
Goddard, M.: Genomic selection: prediction of accuracy and maximisation of long-term response, Genetica, 136, 245–257, https://doi.org/10.1007/s10709-008-9308-0, 2009.
Graffelman, J., Jain, D., and Weir, B.: A genome-wide study of Hardy-Weinberg equilibrium with next generation sequence data, Hum. Genet., 136, 727–741, https://doi.org/10.1007/s00439-017-1786-7, 2017.
Hafid, H., Hasnudi, Bain, H. A., Nasiu, F., Inderawati, Patriani, P., and Ananda, S. H.: Effect of fasting time before slaughtering on body weight loss and carcass percentage of Bali cattle, IOP Conference Series: Earth and Environmental Science, 1–7, Medan, 24–25 October 2019, Medan, Indonesia, https://doi.org/10.1088/1755-1315/260/1/012051, 2019.
Hwang, J. M., Cheong, J. K., Kim, S. S., Jung, B. W., Koh, M. J., Kim, H. C., and Choi, Y. H.: Genetic analysis of ultrasound and carcass measurement traits in a regional Hanwoo steer population, Asian Austral. J. Anim., 27, 457–463, https://doi.org/10.5713/ajas.2013.13543, 2014.
Iwanowska, A., Grześ, B., Mikołajczak, B., Iwańska, E., Juszczuk-Kubiak, E., Rosochacki, S. J., and Pospiech, E.: Impact of polymorphism of the regulatory subunit of the μ-calpain (CAPN1S) on the proteolysis process and meat tenderness of young cattle, Mol. Biol. Rep., 38, 1295–12300, https://doi.org/10.1007/s11033-010-0229-5, 2011.
Jakaria, J., Musyaddad, T., Rahayu, S., Muladno, M., and Sumantri, C.: Diversity of D-loop mitochondrial DNA (mtDNA) sequence in Bali and Sumba Ongole cattle breeds, Journal of the Indonesian Tropical Animal Science, 44, 335–345, https://doi.org/10.14710/jitaa.44.4.335-345, 2019.
Khansefid, M., Pryce, J. P., Bolormaa, S., Chen, Y., Millen, C. A., Chamberlain, A. J., Jagt, C. J. V., and Goddard, E.: Comparing allele specific expression and local expression quantitative trait loci and the influence of gene expression on complex trait variation in cattle, BMC Genom., 19, 793, https://doi.org/10.1186/s12864-018-5181-0, 2018.
Khasrad, Sarbaini, Arfa'i, and Rusdimansyah: Effect of cattle breeds on the meat quality of longissimus dorsi muscles, Pakistan Journal of Nutrition, 16, 164–167, https://doi.org/10.3923/pjn.2017.164.167, 2017.
Li, X., Ekerljung, M., Lundström, K., and Lundén, A.: Association of polymorphisms at DGAT1, leptin, SCD1, CAPN1 and CAST genes with color, marbling and water holding capacity in meat from beef cattle populations in Sweden, Meat Sci., 94, 153–158, https://doi.org/10.1016/j.meatsci.2013.01.010, 2013.
Listyarini, K., Jakaria, Uddin, M. J., Sumantri, C., and Gunawan, A.: Association and expression of CYP2A6 and KIF12 genes related to lamb flavour and odour, Tropical Animal Science Journal, 41, 100–107, https://doi.org/10.5398/tasj.2018.41.2.100, 2018.
Liu, X. D., Moffitt-Hemmer, N. R., Deavila, J. M., Li, A. N., Tian, Q. T., Bravo-Iniguez, A., Chen, Y. T., Zhao, L., Zhu, M. J., Neibergs, J. S., Busboom, J. R., Nelson, M. L., Tibary, A., and Du, M.: Wagyu–Angus cross improves meat tenderness compared to Angus cattle but unaffected by mild protein restriction during late gestation, Animal, 15, 1–7, https://doi.org/10.1016/j.animal.2020.100144, 2021.
Magalhães, A. F. B., Schenkel, F. S., Garcia, D. A., Gordo, D. G. M., Tonussi, R. L., Espigolan, R., Silva, R. M. O., Braz, C. U., Fernandes Júnior, G. A., Baldi, F., Carvalheiro, R., Boligon, A. A., de Oliveira, H. N., Chardulo, L. A. L., and de Alburqurque, L. G.: Genomic selection for meat quality traits in Nelore cattle, Meat Sci., 48, 32–37, https://doi.org/10.1016/j.meatsci.2018.09.010, 2019.
Maltin, C., Balcerzak, D., Tilley, R., and Delday, M.: Determinants od meat quality: tenderness, Proc. Nutr. Soc., 62, 337–347, https://doi.org/10.1079/pns2003248, 2003.
Martojo, H.: Indigenous Bali cattle is most suitable for sustainable small farming in Indonesia, Reprod. Domest. Anim., 47, 10–14, https://doi.org/10.1111/j.1439-0531.2011.01958.x, 2012.
Mateescu, R. G., Garrick, D. J., and Reecy, J. M.: Network analysis reveals putative genes affecting meat quality in Angus cattle, Front. Genet., 8, 171, https://doi.org/10.3389/fgene.2017.00171, 2017.
Nayak, S. N. and Singh V. K.: Marker Assisted Selection, 2nd Edn., Encyclopedia of Applied Plant Science, Whaltham, MA, Academic Press, 183–187, https://doi.org/10.1016/B978-0-12-394807-6.00192-1, 2017.
Nishimura, T.: The role of intramuscular connective tissue in meat texture, Animal Journal Science, 81, 21–27, https://doi.org/10.1111/j.1740-0929.2009.00696.x, 2010.
Park, S. J., Beak, S. H., Jung, D. J. S., Kim, S. Y., Jeong, I. H., Piao, M. Y., Kang, H. J., Fassah, D. M., Na, S. W., Yoo, S. P., and Baik, M.: Genetic, management, and nutritional factors affecting intramuscular fat deposition in beef cattle – A review, Asian Austral. J. Anim., 31, 1043–1061, https://doi.org/10.5713/ajas.18.0310, 2018.
Rego-Costa, A. C., Rosado, E. L., and Soares-Mota, M.: Influence of the dietary intake of medium chain triglycerides on body composition, energy expenditure and satiety: a systematic review, Nutr. Hosp., 27, 103–108, https://doi.org/10.3305/nh.2012.27.1.5369, 2012.
Roopashree, P. G., Shetty, S. S., and Kumari, N. S.: Effect of medium chain fatty acid in human health and disease, Journal of Funtional Foods, 87, 105068, https://doi.org/10.1016/j.jff.2021.104724, 2021.
Rosenvold, K., Peterson, J. S., Laerke, H. N., and Jensen, S. K.: Muscle glycogen stores and meat quality as affected by strategic finishing feeding of slaughter pigs, J. Anim. Sci., 79, 382–391, https://doi.org/10.2527/2001.792382x, 2001.
Santos, C., Moniz, C., Roseiro, C., Tavares, M., Medeiros, V., Afonso, I., Dias, M., and da Ponte, D.: Effects of early post-mortem rate of pH fall and aging on tenderness and water holding capacity of meat from cull dairy holstein-Friesian Cows, J. Food Res., 5, 1–12, https://doi.org/10.5539/jfr.v5n2p1, 2016.
Silva, S. R. and Cadavez, V. P.: Real-Time Ultrasound (RTU) Imaging Methods for Quality Control of Meats, in: Computer Vision Technology in The Food and Beverage Industries, edited by: Sun, D. W., Woodhead Publishing, Cambridge, 277–329, https://doi.org/10.1533/9780857095770.3.277, 2012.
Silver, N., Best, S., Jiang, J., and Thein, S. L.: Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR, BMC Mol. Biol., 7, 33–41, https://doi.org/10.1186/1471-2199-7-33, 2006.
Sinclair, A. J: Docosahexaenoic acid and the brain–What is its role? Asia Pac, Asia Pasific Journal of Clinical Nutrition, 28, 675–688, https://doi.org/10.6133/apjcn.201912_28(4).0002, 2019.
Sirot, V., Oseredzuk, M., Bemrah-Aouachria, N., Volatier, J. L, and Leblanc, J. C.: Lipid and fatty acid composition of fish and seafood consumed in France: CALIPSO study, J. Food Compos. Anal., 21, 8–16, https://doi.org/10.1016/j.jfca.2007.05.006, 2008.
Smith, S. B., Gotoh, T., and Greenwood, P. L.: Current situation and future prospect for global beef production: overview of special issue, Asian Austral. J. Anim., 31, 927–932, https://doi.org/10.5713/ajas.18.0405, 2018.
Sokoła-Wysoczańska, E., Wysoczański, T., Wagner, J., Czyż, K., Bodkowski, R., Lochyński, S., and Patkowska-Sokoła, B.: Polyunsaturated fatty acids and their potential therapeutic role in cardiovascular system disorders, Nutrients, 10, 1561, https://doi.org/10.3390/nu10101561, 2018.
Sun, X., Wu, X., Fan, Y., Mao, Y., Ji, D., Huang, B., and Yang, Z.: Effects of polymorphisms in CAPN1 and CAST genes on meat tenderness of Chinese Simmental cattle, Arch. Anim. Breed., 61, 433–439, https://doi.org/10.5194/aab-61-433-2018, 2018.
Tizioto, P. C., Decker, J. E., Taylor, J. F., Schnabel, R. D., Mudadu, M. A., Silva, F. L., Mourão, G. B., Coutinho, L. L., Tholon, P., Sonstegard, T. S., Rosa, A. N., Alencar, M. M., Tullio, R. R., Medeiros, R. R., Nassu, R. T., Feijo, G. L. D., Silva, L. O. C., Torres, R. A., Siqueira, F., Higa, R. H., and Regitano, L. C. A.: Genome scan for meat quality traits in Nelore beef cattle, Physiol. Genomics, 45, 1012–1020, https://doi.org/10.1152/physiolgenomics.00066.2013, 2013.
Ulum, M. F., Suprapto, E., and Jakaria: Ultrasunograpgy image or Longissimus dorci muscle of Bali cattle, Prosiding KIVNAS PDHI XIII, Konverensi Ilmiah Veteriner Nasional ke-13 Perhimpunan Dokter Hewan Indonesia, 368–369, 23–26 November 2014, Padang, Indonesia, ISBN 978-602-97906-3-4, 2014.
Vallim, T. and Salter, A. M.: Regulation of hepatic gene expression by saturated fatty acid, Prostag. Leukotr. Ess., 82, 211–218, https://doi.org/10.1016/j.plefa.2010.02.016, 2010.
Waples, R. S.: Testing for hardy-weinberg propostions: have we lost the plot?, J. Hered., 106, 1–19, https://doi.org/10.1093/jhered/esu062, 2015.
Whitmore, B. B., Froyen, E., Heskey, C., Parker, T., and Pablo, G. G.: Alpha-linolenic and linoleic fatty acids in the vegan diet: do they require dietary reference intake/adequate intake special consideration?, Nutrients, 11, 2365, https://doi.org/10.3390/nu11102365, 2019.
Xin, J., Zhang, L., Li, Z., Liu, X., Jin, H., and Yan, C.: Association of polymorphisms in the calpain I gene with meat quality traits in Yanbian Yellow Cattle of China, Animal Bioscience, 24, 9–16, https://doi.org/10.5713/ajas.2011.90407, 2011.