the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Molecular characterization, function, tissue differential expression, and single-nucleotide polymorphism of buffalo TP53 gene
Lige Huang
Dan Sheng
Xinyang Fan
Ruixia Gao
Yongwang Miao
TP53 has been shown to be involved in lactation in cattle. However, the role of TP53 in buffalo lactation remains unknown. To this end, we isolated and identified the complete coding sequence (CDS) of the TP53 gene from the buffalo mammary gland and further analyzed its molecular characteristics, function, tissue differential expression, and single-nucleotide polymorphism (SNP). A transcript of this gene was cloned with a CDS length of 1161 bp, encoding a protein consisting of 386 amino acid residues. Bioinformatics analysis showed that buffalo TP53 CDS and the physicochemical characteristics, conserved domains, structure, and function of its encoded protein are highly similar to those of other species in Bovidae. The buffalo TP53 protein contains an N-terminal activation domain, a DNA-binding domain, and a tetrameric domain, and it plays a functional role in the nucleus. TP53 was found to express in all 11 detected buffalo tissues, and its expression in the heart, kidney, brain, muscle, and rumen during lactation was significantly higher than that during non-lactation (p<0.05), while in the liver, lung, and mammary gland, its expression was the opposite (p<0.05). Interference experiments in buffalo mammary epithelial cells (BuMECs) showed that TP53 inhibits the expression of genes related to milk protein and milk fat synthesis through the PI3K–AKT–mTOR pathway. A synonymous nucleotide substitution (c.204C > T) was found in the TP53 CDS of river buffalo, which is the CC homozygote in swamp buffalo. The results indicate that the TP53 gene is involved in buffalo lactation by negatively regulating the synthesis of milk protein and milk fat.
- Article
(6097 KB) - Full-text XML
-
Supplement
(1213 KB) - BibTeX
- EndNote
The TP53 (tumor protein 53, TP53/P53) gene is the tumor suppressor gene with the highest mutation frequency in tumorgenesis, and about 50 % of human cancers are associated with TP53 mutations (Vogelstein et al., 2000). As a transcription factor, TP53 trans-activates the transcription of downstream target genes by specifically binding to their regulatory sequences in response to a variety of cellular stresses, thus participating in a variety of biological processes such as cell cycle control, apoptosis, aging, differentiation, and DNA repair (Levine and Oren, 2009). In addition, the activation of TP53 in human lung cancer cells leads to dephosphorylation of the eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1), which increases the binding of eukaryotic initiation factor 4E (EIF4E) to 4EBP1 and then inhibits protein synthesis (Tilleray et al., 2006). In mouse erythroleukemia cells, the activation of TP53 decreased the phosphorylation of 4EBP1 and the activity of ribosomal protein S6 kinase (S6k), resulting in downregulation of protein synthesis (Horton et al., 2002). With further research, TP53 has been revealed to be involved in cellular glycolysis, oxidative phosphorylation, amino acid metabolism, and other metabolic pathways (Maddocks and Vousden, 2011). Subsequently, TP53 has been proven to be involved in regulating the expression of genes related to lipid metabolism, thereby promoting fatty acid oxidation and inhibiting lipid synthesis (Goldstein and Rotter, 2012). In mouse 3T3-L1 preadipocytes, the knockout of the TP53 gene increases fat synthesis and adipogenic gene expression, while its overexpression results in the opposite (Okita et al., 2014). The TP53 signaling pathway is also significantly correlated with the milk yield, protein yield, and lactose rate of Holstein cows (Ammah et al., 2018).
The buffalo TP53 gene is located on chromosome 3, and the NCBI gene database (https://www.ncbi.nlm.nih.gov/, last access: 4 February 2024) shows that there are three transcript variants in the mRNA sequences of the buffalo gene. The transcript variant X1 (NM_001290844) contains 10 exons, with a complete coding sequence (CDS) of 1161 bp in length, which encodes a polypeptide containing 386 amino acid residues, while transcript variants X2 (XM_006062871) and X3 (XM_044938378) were predicted from the buffalo genomic data, and both of them contain 11 exons (https://www.ncbi.nlm.nih.gov/, last access: 4 February 2024). It has been determined that buffalo TP53 protein plays a functional role in the nucleus (Singh et al., 2015). In addition, buffalo TP53 protein was found to efficiently transcribe and activate the cyclin-dependent kinase inhibitor 1A (CDKN1A) in buffalo (Singh et al., 2015). In cattle, the TP53 gene is located on chromosome 19, containing 11 exons and 10 introns, and its CDS (NM_174201) length is also 1161 bp (https://www.ncbi.nlm.nih.gov/, last access: 4 February 2024).
Buffalo are divided into two distinct subtypes: river buffalo (2n=50) and swamp buffalo (2n=48) (Shi et al., 2012). River buffalo are used primarily for milk production, and swamp buffalo are primarily used for draft. Buffalo milk is the second largest source of milk around the world, accounting for 13 % of the world's total milk production (Nasr et al., 2016). China is the third largest country in the world with respect to buffalo farming, after India and Pakistan (Abdel Hamid et al., 2023), and the Binglangjiang buffalo is a river-type buffalo that has been found in western Yunnan, China, in recent years. Compared with cow's milk, buffalo milk is rich in protein (especially casein), fat, and lactose as well as minerals, such as calcium, magnesium, and inorganic phosphate (Ahmad et al., 2008). So far, it has been found that TP53 plays an important role in the metabolic regulation of some mammals, but its functional role in buffalo, particularly in lactation, remains unclear. The aim of this study is to isolate and identify the buffalo TP53 gene from the tissue of the buffalo mammary gland and to further describe its molecular characteristics, function, tissue differential expression, interaction, and single-nucleotide polymorphism (SNP) through bioinformatics, comparative genomic methods, and gene function experiments.
2.1 Sample collection
Six Binglangjiang buffalo with their third calf aged about 4 years were selected for sample collection, including three in the peak lactation period (about 60 d postpartum) and three in the dry lactation period (about 60 d before parturition). All the buffalo sampled were fed and managed under the same conditions. After the buffalo were slaughtered, the tissue samples used to extract total RNA were collected immediately from the heart, liver, spleen, lung, kidney, brain, cerebellum, small intestine, muscle, mammary gland, and rumen, and then we put them into RNase-free freezing tubes, stored them in liquid nitrogen, and then took them back to the laboratory for total RNA preparation.
The blood samples used for SNP detection of TP53 CDS included those of 60 Binglangjiang buffalo (river type) and those of 60 Dehong buffalo (swamp type). The buffalo were all adults and were not directly related. The Binglangjiang buffalo and Dehong buffalo were sampled from their core distribution areas, Tengchong and Mangshi in Yunnan Province, China, respectively. Approximately 4 mL of blood sample was collected from each buffalo in centrifuge tubes containing EDTA anticoagulant and were stored in the laboratory at low temperatures for DNA extraction.
2.2 Isolation and identification of TP53
Total RNA was extracted using the RNAiso Plus kit (TaKaRa, Dalian, China) according to the merchant product manual. The concentration and purity of RNA were detected by an ultraviolet spectrophotometer (Thermo Fisher Scientific, CA, USA), and then the integrity of the RNA was assessed through 1 % agarose gel electrophoresis. The cDNA was synthesized from 2–3 µg RNA using the Reverse Transcription Kit (TaKaRa, Dalian, China) and Oligo(dT)18 (50 µM; TaKaRa, Dalian, China) as the primer. Then, the cDNA was diluted to 100 ng mL−1 and kept in a refrigerator at −20 °C.
Primers for isolating the CDS of buffalo TP53 were designed using the mRNA sequence (XM_006062871) of buffalo TP53 as the template (Table S1 in the Supplement). The total volume of the PCR reaction system was 10 µL, which contained 5 µL of 2× Es Taq Master Mix (CWBIO, Beijing, China), 0.5 µL (each) of forward and reverse primers (10 µmol L−1), 1 µL of the template (100 ng µL−1), and 3 µL of ddH2O. The PCR was conducted under pre-denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 63.9 °C for 40 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min, and then it was stopped at 4 °C. PCR products were detected by 1 % agarose gel electrophoresis, and the target band was cut and further purified using the TIANgel Purification Kit (TIANGEN, Beijing, China). The purified PCR products were linked to the pMD-18T vector (TaKaRa, Dalian, China) and performed molecular cloning. A total of 60 clones were selected and sequenced bidirectionally.
The obtained sequence of buffalo TP53 was checked and proofread using the Lasergene software package (DNASTAR Inc.), and the open reading frame (ORF) was identified by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/, last access: 5 March 2023). In order to confirm that the obtained sequence is the target sequence, a homologous search was conducted in the NCBI database, using it as the query sequence, by means of the online BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi, last access: 5 March 2023).
2.3 Molecular characteristics and function prediction
TP53 sequences of 13 species were recruited from the NCBI database for the analysis of sequence identity, gene structure, motif, and conserved domain (Table S2). The TP53 structure information of the species of Bovidae, human, rat, horse, deer, camel, and pig was derived from GTF files downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/datasets/, last access: 6 March 2023). The files of each species were further processed by TBtools, and the transcriptional region structure of TP53, including untranslated regions and exon–intron structure for each species, was visualized by the online Gene Structure Display Server (http://gsds.gao-lab.org/, last access: 6 March 2023). The nucleotide sequence consistency among multiple species was estimated by MegAlign (DNASTAR Inc.). Based on Jones–Taylor–Thornton and gamma-distributed models, the phylogenetic analysis of TP53 protein sequences was constructed using MEGA7 (Kumar et al., 2016). The motif composition for TP53 was obtained by submitting its amino acid sequences to the MEME Suite website (https://meme-suite.org/meme/, last access: 6 March 2023), and the conserved structural domain was predicted by submitting its protein sequences to the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/?term=, last access: 6 March 2023) in the NCBI. The functional modification site, physical and chemical characteristics, hydrophilicity, signal peptide, and transmembrane domain of TP53 were predicted using the online software PROSITE (http://prosite.expasy.org/prosite.html, last access: 6 March 2023), ProtParam (https://web.expasy.org/protparam/, last access: 6 March 2023), ProtScale (https://web.expasy.org/protscale/, last access: 6 March 2023), the SignalP5.0 server (https://services.healthtech.dtu.dk/services/SignalP-5.0/, last access: 6 March 2023), and the TMHMM 2.0 server (https://services.healthtech.dtu.dk/services/TMHMM-2.0/, last access: 6 March 2023), respectively. The subcellular localization and the secondary and three-dimensional structures of the protein were determined by the online ProtComp (http://linux1.softberry.com/berry.phtml, last access: 6 March 2023), SOPMA (http://npsa-pbil.ibcp.fr/, last access: 6 March 2023), and SWISS-MODEL (http://swissmodel.expasy.org/, last access: 6 March 2023), respectively. The molecular function and biological pathway of the protein were analyzed using InterPro (http://www.ebi.ac.uk/interpro/, last access: 6 March 2023), UniProt (http://www.uniprot.org/, last access: 6 March 2023), and DAVID (http://david.ncifcrf.gov/, last access: 6 March 2023). Protein–protein interactions between the target protein and other proteins were predicted by the online STRING (https://string-db.org/, last access: 6 March 2023).
2.4 Cell culture
Buffalo mammary epithelial cells (BuMECs) were obtained from the mammary tissue of lactating buffalo (60 d postpartum), as previously described in our laboratory (Fan et al., 2020). Purified and identified BuMECs were cultured in Dulbecco's modified Eagle medium (DMEM) containing 5 µg mL−1 hydrocortisone (Sigma, St. Louis, MO, USA), 5 µg mL−1 insulin (Sigma, St. Louis, MO, USA), 1 µg mL−1 epidermal growth factor (Sigma, St. Louis, MO, USA), 2 % penicillin–streptomycin (Gibco, Carlsbad, CA, USA), and 10 % fetal bovine serum (Gibco, Carlsbad, CA, USA) and were incubated under the conditions of 37 °C and 5 % CO2. The medium was changed every 48 h. Before the beginning of experiments, BuMECs were cultured for 48 h in the above medium with 3 µg mL−1 prolactin (Sigma, St. Louis, MO, USA) to induce lactogenesis. Finally, the medium was replaced with hormone- and growth-factor-free DMEM supplemented with 10 % fetal bovine serum.
2.5 Transfection of small interfering RNAs
Two specific small interfering RNAs (siRNAs) targeting different regions of TP53 CDS (siRNA1-TP53 and siRNA2-TP53) and one non-specific negative control siRNA (NC) were designed and synthesized by Shanghai Biological Engineering Technology Services Co., Ltd. The TP53 and NC siRNA sequences are presented in Table S1 in the Supplement. The interference efficiency of siRNA-TP53 was assessed by comparing the mRNA abundance of TP53 in BuMECs transfected with siRNA-TP53 and transfected with NC. The expression level of TP53 was detected by qPCR for 48 h after transfection. When the confluence of cells reached 70 %–80 %, siRNA (100 nM) with the best interference efficiency was then transfected into BuMECs using TransLipid Transfection Reagent (TransGen Biotech, Beijing, China) according to the manufacturer's instructions. Cells were harvested 48 h after transfection for RNA extraction, with three replicates for each group.
2.6 Detection and analysis of gene expression
The differential expression of TP53 in lactating and non-lactating buffalo tissues and the expression of the genes related to milk protein and milk fat synthesis in BuMECs were detected by qPCR. The ubiquitously expressed housekeeping genes ACTB, GAPDH, and RPS23 were used as internal references to normalize the results of the target gene. Primers used for qPCR detection are listed in Table S1. The qPCR was performed using the SYBR Premix Ex Taq fluorescent dye (TaKaRa, Dalian, China) and the iQ5 RT-PCR instrument (Bio-Rad, CA, USA). The total volume of qPCR was 20 µL, which contained 10 µL of SYBR Premix Ex Taq, 0.8 µL (each) of upstream and downstream primer (10 µM), 6.4 µL of ddH2O, and 2 µL of cDNA (100 ng µL−1). The reaction procedure was conducted according to the manufacturer's instructions. The program LinRegPCR was used to determine the qPCR amplification efficiency of TP53. All analyses were performed in triplicate.
Results were statistically analyzed and visualized using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA), and all experimental data were expressed as the means ± standard error of the means (mean ± SEM) from each of the three independent experiments. qPCR data were analyzed using the 2−ΔΔCt method relative to the control. Statistical analysis was performed using Student's t test, and either p<0.05 (significant) or p<0.01 (highly significant) was considered to be statistically significant.
2.7 Detection and analysis of polymorphisms
Genomic DNA was extracted from the blood sample using the method of phenol–chloroform extraction (Sambrock and Russell, 2001). The variation in the TP53 CDS was detected by direct bidirectional sequencing of PCR products. Primers for SNP detection were designed based on the sequence of buffalo TP53 (NC_059159; Table S1).
The PCR system contained 5 µL of 2× Es Taq Master Mix (CWBIO, Beijing, China), 0.5 µL (each) of upstream and downstream primers (10 µmol L−1), 1 µL of DNA template (100 ng µL−1), and 3 µL of ddH2O. The PCR was performed with a pre-denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at a specific temperature (Table S1) for 40 s, extension for a specific time (Table S1) at 72 °C, and further extension at 72 °C for 5 min, finally stopping the reaction at 4 °C. The PCR products were detected by 1 % agarose gel electrophoresis; bands were then cut, and we purified them by means of the TIANgel Purification Kit (TIANGEN, Beijing, China). The purified PCR products were bidirectionally sequenced by Shanghai Biological Engineering Technology Services Co., Ltd (Shanghai, China).
The sequences were first checked and proofread and then were output by the SeqMan program (DNASTAR Inc.) and MEGA7 (Kumar et al., 2016) to obtain the information of SNPs. The relative synonymous codon usage (RSCU) for CDS was analyzed using the CodonW (http://codonw.sourceforge.net/, last access: 6 February 2024) program. Multiple nucleotide sequences were compared using MegAlign (DNASTAR Inc.), Clustal X (Larkin et al., 2007), and BioEdit (Hall, 1999) software, and nucleotide and amino acid difference sites were output using MEGA7 software (Kumar et al., 2016).
3.1 Isolation and identification of buffalo TP53 gene
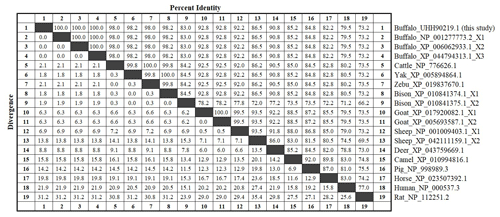
In this study, only one type of transcript was obtained from the sequencing results of the 60 monoclonal colonies selected (Fig. S1 in the Supplement). The CDS of this gene was determined to be 1161 bp in length by the ORF Finder program, and the CDS was submitted to the NCBI database under the accession number of OL456217. Then the homologous search in the NCBI database using the CDS as the query sequence showed that the consistency of it with the TP53 CDS of buffalo (NM_001290844.2), cattle (NM_174201.1), yak (XM_005894802.2), zebu (XM_019982111.1), bison (XM_010843072.1), bison (XM_010843073.1), sheep (NM_001009403.1), sheep (XM_042255225.1), goat (XM_005693530.3), and goat (XM_018064593.1) is 99.7 %, 98.3 %, 98.4 %, 98.3 %, 98.4 %, 98.3 %, 96.0 %, 93.0 %, 96.2 %, and 96.2 %, respectively (Fig. S2). Therefore, it was identified as the buffalo TP53. The composition of A, T, G, and C of buffalo TP53 CDS is 23.08 %, 25.41 %, 20.67 %, and 30.84 %, respectively, and the content of G + C is 56.26 %. Buffalo TP53 encodes a protein composed of 386 amino acid residues (Fig. 1).
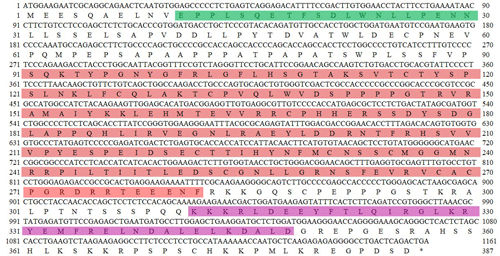
Figure 1The CDS of buffalo TP53 and its encoded amino acid sequence in this study. ATG – start codon; * – stop codon. The region in green represents the N-terminal activation domain (TAD), the region in red represents the DNA-binding domain (DBD), and the region in purple represents the tetrameric domain (TD).
3.2 Structure of transcriptional region and consistency of amino acid sequence
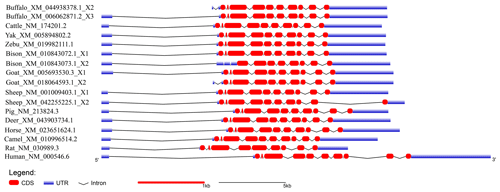
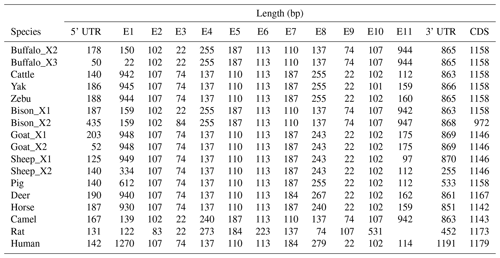
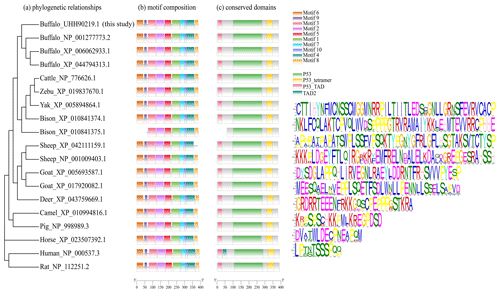
In order to demonstrate the structural characteristics of buffalo TP53, the transcriptional region of this gene was compared with the homologous sequence of the livestock in Bovidae and other mammals (Fig. 2 and Table 1). Until now, there have been three buffalo TP53 mRNA sequences in the NCBI database, but only two of them contain flanking sequences, and they were all included in the analysis of this study. The two buffalo transcript variants have the same CDS length but differ with respect to the 5' flanking sequences. The length and structure of buffalo TP53 CDS are nearly identical to those of cattle, yak, zebu, bison, and goat, except for the fact that the bison TP53 transcript variant X2 has a shorter CDS than that of buffalo, and the coding initiation site of the bison TP53 variant X2 is located at exon 4. There are two transcriptional variants of TP53 in sheep, and their differences are reflected in the different positions of exon 11 in the transcriptional variants X1 and X2, which are located at positions 11 971–12 919 and 16 195–16 528 of the genome sequence (NC_056064), respectively, in accordance with the GU-AG rule for intron splicing. The sequences of the 5' flanking region and 3' flanking region for buffalo TP53 are different from those of other species in Bovidae. The TP53 of all species in Bovidae contains 11 exons and 10 introns.
The amino acid sequences of buffalo TP53 obtained in this study were aligned with those of some species recruited from the NCBI database (Fig. 3). The results showed that the sequence identity of TP53 from buffalo in this study is 83.0 % and 86.5 % with the TP53 transcript variant X2 from bison and sheep, respectively, and ranged from 92.2 % to 100 % with TP53 sequences from six other species of Bovidae.
3.3 Physicochemical characteristics and functional modification sites
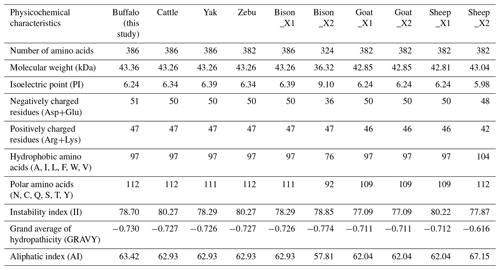
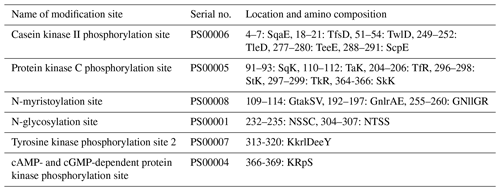
The physicochemical characteristics of buffalo TP53 were analyzed using the transcript variants of other species in Bovidae as controls, and the results showed that the basic physicochemical characteristics of buffalo TP53 are very similar to those of other species in Bovidae, except for the basic physicochemical characteristics of the TP53 transcript variant X2 in bison and sheep (Table 2). The grand average of the hydrophobicity of the buffalo TP53 protein is −0.730, indicating that the protein is hydrophilic. Prediction showed that the buffalo TP53 protein has no signal peptide and transmembrane helix, and there are six potential functional modification sites harbored in it (Table 3).
3.4 Motifs, conserved domains, and evolutionary relationship
The motif composition and conserved domains of multiple mammalian TP53 proteins were analyzed, and a total of 10 motifs and 4 conserved domains were identified (Fig. 4). The DBD (AA101-282) of buffalo TP53 is composed of motifs 1, 2, 4, 5, and 7. The TD (AA312-348) is composed of motif 3, and the TAD (AA7-30) is composed of motif 6. The protein encoded by the transcript variant X2 in bison lacks motif 6, motif 9, and domain TAD. The TP53 protein lacking the domain TAD can inhibit the transcriptional activation of the normal TP53 protein, mediate the export of the intact TP53 from the nucleus to the cytoplasm, and undergo monoubiquitination in an MDM2-independent manner (Ghosh et al., 2004). The transcription variant X2 of sheep TP53 lacks motif 8 but does not affect the conserved domain composition of the protein. In human, the TAD is composed of TAD1 and TAD2 (Raj and Attardi, 2017). A phylogenetic tree constructed based on TP53 amino acid sequences with rat and human as the outgroup is shown in Fig. 4. The results show that buffalo are clustered with the animals of the Bos genus, suggesting that buffalo TP53 has a similar function to that of such animals.
3.5 Secondary and three-dimensional structure
The secondary structure composition of buffalo TP53 is highly similar to that of other species in Bovidae, except that of the TP53 transcript variant X2 in bison and sheep (Fig. S3 and Table S3). The predicted three-dimensional structure of buffalo TP53 is highly similar to that of other species in Bovidae, except that of the bison TP53 transcript variant X2 (Fig. S4).
3.6 Proteins interacting with buffalo TP53
Ten proteins were predicted to interact with buffalo TP53 (Fig. 5). There are three kinds of proteins in the network: one is related to lipid synthesis, which includes MDM2 (murine double minute 2), CDKN1A, and EP300 (E1A-binding protein P300); the other is related to protein synthesis, which includes ATM (ataxia–telangiectasia mutated) and SIRT1 (silent information regulator 1); and another is associated with apoptosis, which includes MAPK8 (mitogen-activated protein kinase 8), MDM4 (murine double minute 4), USP7 (ubiquitin-specific peptidase 7), and CHEK1 (checkpoint kinase 1).
3.7 Subcellular localization, biological processes, and molecular function
The buffalo and cattle TP53 proteins were predicted to be localized in the nucleus (100 %) and involved in the biological processes of apoptosis (GO: 0006915), biological rhythms (GO: 0048511), cell cycle (GO: 0007049), transcription, transcription regulation, and necrosis. They are mainly involved in the molecular functions of DNA-binding transcription factor activity (GO: 0003700), DNA binding (GO: 0003677), transcription cis-regulatory region binding (GO: 0000976), and metal ion binding (GO: 0046872). In addition, they participate in the MAPK signal pathway, TP53 signal pathway, PI3K–AKT signal pathway, cell cycle, apoptosis, and cell senescence.
3.8 Tissue differential expression
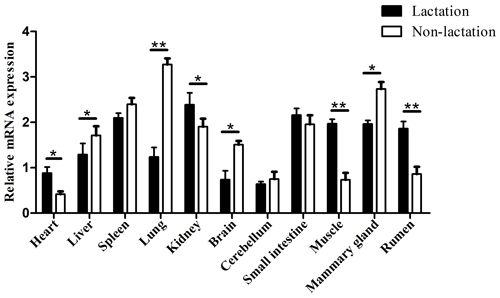
To ensure that the results of the qPCR detection were the target gene, all amplification products were verified by sequencing and BLAST alignments (Table S4). Among the 11 buffalo tissues of lactation and non-lactation detected, TP53 was found to express in all tissues (Fig. 6). In the non-lactation period, its expression levels were relatively high in the lung, mammary gland, and spleen, while in the lactation period, its expression was relatively high in the kidney, small intestine, spleen, muscle, mammary gland, and rumen. It is worth noting that the expression of TP53 in the heart, kidney, brain, muscle, and rumen during lactation was significantly higher than that during non-lactation (p<0.05), while its expression in the liver, lung, and mammary gland was just the opposite (p<0.05).
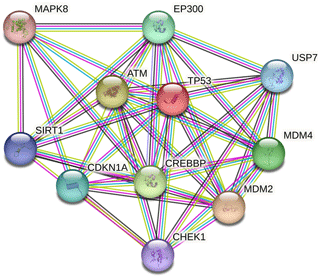
Figure 5Protein–protein interaction network of buffalo TP53. Several putative interactors were predicted for TP53 protein. The colors of the lines represent the different modes by which the interaction was predicted. Light-green color is for text mining, red line color is for gene fusions, purple color is for experimentally determined, and cyan color indicates attainment from curated databases.
3.9 Knockdown of buffalo TP53 promotes the synthesis of milk protein and milk fat in BuMECs
Here, the interference efficiency of two specific siRNAs targeting the TP53 CDS was evaluated in BuMECs. Compared with negative control (siRNA-NC)-treated cells, the mRNA abundance of this gene was significantly reduced in cells transfected with specific siRNAs, with an 87 % reduction for siRNA1-TP53 and a 69 % reduction for siRNA2-TP53 (p<0.05; Fig. S5). Therefore, we transfected siRNA1-TP53 into BuMECs for further functional experiments due to its higher interference efficiency.
The mRNA abundance of TP53 in lactating and non-lactating BuMECs was examined by qPCR. The results showed that the mRNA abundance of TP53 in BuMECs was significantly lower during lactation than during non-lactation (p<0.05; Fig. 7). In order to determine whether TP53 expression is involved in the synthesis of buffalo milk protein and milk fat, we performed knockdown experiments on TP53 by transfecting siRNA1-TP53 into BuMECs.
We successfully knocked down the TP53 gene in BuMECs and further detected changes in the expression levels of genes related to milk protein and milk fat synthesis in BuMECs under TP53 gene knockdown. The knockdown of TP53 led to changes in the expression of the PI3K, AKT1, mTOR, S6K1, EIF4E, and CSN2 genes related to milk protein synthesis, as well as in the SREBF1, PPARG, ACACA, FASN, and SCD genes related to milk fat synthesis. In comparison to the group of negative control (siRNA-NC), the expressions of the PI3K, AKT1, mTOR, S6K1, EIF4E, and CSN2 genes were increased 1.85-fold (p<0.05), 0.93-fold (p<0.05), 1.23-fold (p<0.01), 0.80-fold, 1.10-fold, and 1.17-fold (p<0.05), respectively, in the group treated with siRNA1-TP53 (Fig. 8a). In addition, the SREBF1, PPARG, ACACA, FASN, and SCD genes increased 1.61-fold (p<0.01), 0.83-fold (p<0.05), 1.12-fold (p<0.05), 1.34-fold (p<0.05), and 0.76-fold, respectively (Fig. 8b).
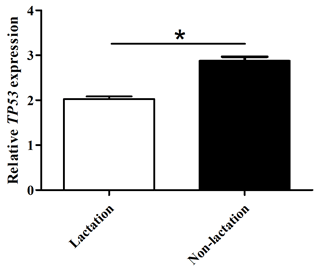
Figure 7The mRNA abundance of the TP53 gene in lactating and non-lactating BuMECs. The values are presented as means ± SEM; * – p<0.05; – p<0.01.
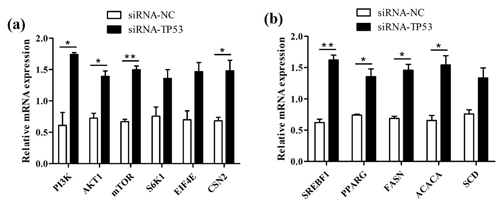
Figure 8The effects of TP53 knockdown on genes related to milk protein and milk fat synthesis in BuMECs. (a) The effect of TP53 knockdown on the expression of genes related to milk protein synthesis. (b) The effect of TP53 knockdown on the expression of genes related to milk fat synthesis. The values are presented as means ± SEM; * – p<0.05; – p<0.01.
3.10 Population variation
In this study, one synonymous substitution was found in the TP53 CDS of river buffalo, that is, c.204C > T, which had three genotypes of CC, CT, and TT, with a high frequency of C allele, while the CC homozygote was found at c.204 in swamp buffalo. Codon analysis of the TP53 gene showed little difference in codon preference between the sequence (B1, this study) of c.204C and the sequences (XM_006062871 and NM_001290844 from the NCBI database, named B2 and B3, respectively) of c.204T (Table S5). The sequence B3 was found to have two nucleotide differences compared to the sequences B1 and B2 at c.12 and c.117 (Fig. 9).
3.11 Nucleotide sequence differences in Bovidae species
The sequences of buffalo TP53 in this study were aligned with the TP53 sequences of Bovidae species downloaded from the NCBI database (Fig. 9 and Table S6; only one sequence was taken if they were identical). There are 17 nucleotide difference sites between buffalo and other species of Bovidae, which are c.111, c.179, c.204, c.258, c.339, c.588, c.678, c.684, c.705, c.798, c.844, c.883, c.932, c.944, c.953, c.960, and c.982. Among them, six nucleotide sites of c.179, c.844, c.883, c.944, c.953, and c.982 lead to the amino acid differences of TP53 protein sequences (Fig. S6). It is noteworthy that there is an InDel (insertion–deletion) codon in the TP53 gene between buffalo, the Bos genus, and the Ovis genus.

Figure 9Differential sites for TP53 nucleotide sequences between buffalo and other species of Bovidae. Numbers indicate the position of the coding region. Point (⋅) indicates consistency with the sequence B1. Nucleotide substitutions are represented by different letters. Missing information is marked with a question mark (?). A horizontal line (–) indicates a deletion in the sequence.
In this study, the complete coding sequence of the buffalo TP53 gene was isolated and identified. An mRNA transcript of buffalo TP53 was obtained, and its CDS encodes a polypeptide consisting of 386 amino acid residues, which is consistent with a previous report (Singh et al., 2015). The TP53 protein of buffalo is a stable hydrophilic protein without a signal peptide and transmembrane domain, which plays a transcriptional regulatory role in the nucleus as a transcription factor. The alignment of sequences revealed that, except for the molecular characteristics of the TP53 transcript variant X2 in bison and sheep, the structure of the transcriptional region of TP53, the basic physicochemical characteristics, secondary structure composition, tertiary structure, motif composition, and conserved domains of the TP53 protein in species of Bovidae are highly consistent, although there are some interspecies differences. Phylogenetic analysis showed that buffalo and the livestock in the Bos genus were clustered together, which indicates that the function of the buffalo TP53 is more similar to those of animals in the Bos genus. Previous study has found that mouse TP53 contains three domains of TAD, DBD, and TD (Lane, 1992), which are shared by the members of the TP53 family (Kaelin, 1999). The TAD domain is required for TP53 to achieve transcriptional activation activity and can interact with a variety of proteins, including the transcription factors TFIID and TFIIH, as well as several TATA box-binding protein-associated factors (TAFs) (Kamada et al., 2016). The TD domain can regulate the TP53 oligomeric state and DNA-binding activity, and the DBD enables TP53 to bind to relevant DNA sequences to play a functional role (Kamada et al., 2016). In this study, we found that buffalo TP53 also possesses the three conserved domains that are consistent with those found in mice. Comparative analysis suggests that the function of TP53 is conservative. It is functionally consistent in mammals, and that of buffalo is particularly consistent with that of the species of the Bos genus.
The post-translational modification of the TP53 protein is an important mechanism by which TP53 responds to a variety of signal stimuli, including phosphorylation, ubiquitination, acetylation, and methylation (Kruse and Gu, 2008). Phosphorylation modification of TP53 is a key step to stabilize TP53 and enhance TP53 transcriptional activity (Nguyen et al., 2014). Activation of PKC by genotoxic stress induces phosphorylation of TP53 on Ser46, resulting in the commitment of cell fate to apoptosis (Yoshida et al., 2006). The casein kinase 2 (CK2) can phosphorylate the C-terminal Ser392 of TP53, resulting in an increase in the specific binding of TP53 to DNA (Leblanc and May, 2002). In this study, we found that buffalo TP53 also has phosphorylation sites of PKC and CK2, but whether these sites play a functional role in the post-transcriptional regulation of buffalo TP53 remains to be further investigated.
Previous study in mice has found that TP53 is highly expressed only in the brain, liver, lung, intestine, kidney, thymus, and salivary glands during the differentiation stage of various organ tissues but not in other organs and tissues (Schmid et al., 1991). In this study, the differential expression analysis of TP53 displayed that TP53 was expressed in all 11 buffalo tissues during lactation and non-lactation, which indicates that this gene plays a functional role in multiple tissues of buffalo. The expression of buffalo TP53 in the heart, kidney, brain, muscle, and rumen during lactation was significantly higher than that during non-lactation, while the expression of TP53 in the liver, lung, and mammary gland was the opposite, which might be related to the different physiological states of buffalo. It is worth noting that the mRNA abundance of TP53 in BuMECs was also lower during lactation than during non-lactation. It has been shown that the knockdown of TP53 leads to delayed degeneration of mammary epithelial cells in mice (Jerry et al., 1998). Therefore, it can be inferred that buffalo TP53 is related to lactation.
In this study, buffalo TP53 was predicted to interact with ATM and SIRT1. Study has reported that cytoplasmic ATM can regulate protein synthesis in the insulin signaling pathway by phosphorylating 4E-BP1 (Yang and Kastan, 2000). SIRT1 negatively regulates the mammalian target of rapamycin (mTOR) signaling through the tuberous sclerosis complexes 1 and 2 (TSC1 and TSC2) (Ghosh et al., 2010). It is therefore hypothesized that buffalo TP53 is involved in protein synthesis. In mammals, milk protein synthesis is regulated by the expression of genes downstream of the mTOR pathway associated with milk protein synthesis, such as S6K1 and eIF4E (Rius et al., 2010; Wang et al., 2006). The eIF4E and S6K1 genes play an essential role in the translation of ribosomal mRNA sequences, and mTOR ultimately participates in protein synthesis by affecting their expression levels (Hayashi et al., 2009; Saxton and Sabatini, 2017). β-casein (CSN2) is the second most abundant protein in cows' milk and can be used as an indicator of protein synthesis ability in bovine mammary epithelial cells (BMECs) (Chatchatee et al., 2001). However, TP53 can induce the expression of genes that negatively regulate the insulin-like growth factor 1 (IGF1)–protein kinase B (AKT) pathway and mTOR pathway, which include phosphatase and tension homolog (PTEN), TSC, and AMP-activated protein kinase (AMPK) genes (Zhang et al., 2010). In this study, the mRNA abundance of phosphoinositide 3-kinase (PI3K), AKT1, mTOR, S6K1, EIF4E, and CSN2 genes associated with milk protein synthesis were increased after interfering with TP53 in BuMECs. It has been reported that TP53 inhibits the synthesis of milk protein through the mTOR pathway in the lactating mammary gland of dairy cows (Bionaz and Loor, 2011). Therefore, the results can indicate that buffalo TP53 inhibits milk protein synthesis.
Proteins predicted to interact with buffalo TP53 include MDM2, CDKN1A, EP300, SIRT1, and ATM in this study. MDM2 is an E3 ubiquitin ligase with carcinogenic effect, which can promote adipogenesis by enhancing CREB-mediated trans-activation and limiting TP53 activity (Hallenborg et al., 2012). TP53 was found to induce the expression of CDKN1A, which promotes lipid accumulation in mouse liver and white adipose tissue (Takasaki et al., 2012) and leads to an increase in the apoptosis threshold in the cells of human lung cancer (Marijn et al., 2019). EP300 (P300) is an acetyltransferase that transcriptionality upregulates the expression of fatty acid synthase (FASN) and promotes the accumulation of lipids (Gang et al., 2016). SIRT1 regulates lipid homeostasis in mouse liver by positively regulating the peroxisome proliferator-activated receptor α (PPARα) (Purushotham et al., 2009). The ATM gene encodes a serine–threonine protein kinase of the phosphatidylinositol-3 kinase-related protein kinase (PIKK) superfamily (Khalil et al., 2012), which has been reported to be highly expressed in well-differentiated adipocytes in pigs, suggesting that it may participate in adipocyte differentiation (Wang and Proud, 2006). It is therefore hypothesized that buffalo TP53 is involved in the process of fat metabolism. In mammary tissues, sterol regulatory element-binding proteins (SREBPs) and PPARγ are important transcriptional regulators in the fatty acid synthesis gene network, directly regulating genes such as FASN, acetyl-CoA carboxylase alpha (ACACA), and stearoyl-CoA desaturase (SCD) that affect milk fat transport and synthesis (Carreño et al., 2016; Liu et al., 2016). FASN, SCD, and ACACA are central enzymes for milk fat synthesis. In this study, the mRNA abundance of the SREBF1, PPARG, ACACA, FASN, and SCD genes involved in milk lipid synthesis were elevated after knockdown of TP53 in BuMECs. Previous study showed that TP53 inhibits the expression of SREBF1 and FASN genes in the adipocytes of mice, thereby inhibiting the synthesis of fat (Yahagi et al., 2003). Therefore, the results showed that TP53 inhibited the synthesis of milk fat in buffalo. This study predicts that buffalo TP53 participates in the PI3K–AKT pathway. This pathway is the upstream of the mTOR pathway and plays a crucial role in protein and lipid synthesis (Appuhamy, 2010; Smith et al., 2008). Studies have also found that the mTOR pathway can affect the expression of SREBPs and PPARγ, thereby regulating fatty acid synthesis (Li et al., 2017; Huang et al., 2020; Caron et al., 2015). These results indicate that TP53 negatively regulates the synthesis of milk protein and milk fat in BuMECs through the PI3K–AKT–mTOR pathway.
In mice, a study showed that the substitution of arginine for leucine at the position 172 of TP53 in the mammary gland can lead to lactation failure (Li et al., 1994). In this study, an SNP (c.204C > T) was found in the coding region of TP53 in river buffalo, where the nucleotide substitution is synonymous and does not affect the preference of codon usage; thus, it is presumed to have no effect on the function of buffalo TP53. In addition, there are 17 nucleotide difference sites in CDS between buffalo and other species in Bovidae, 6 of which lead to amino acid differences. These sites can be used as molecular markers to distinguish buffalo from other species of Bovidae.
The molecular characteristics and function of the buffalo TP53 gene and its encoded protein are highly similar to those of other species in Bovidae, indicating that it is functionally conserved. It plays a functional role as a transcription factor in the nucleus and is involved in metabolic regulation related to the PI3K–AKT pathway. TP53 plays an important functional role in multiple tissues of buffalo and negatively regulates milk protein and milk fat synthesis in the tissue of the mammary gland through the PI3K–AKT–mTOR pathway. The synonymous substitution (c.204C > T) found here in river buffalo has limited potential value in buffalo-breeding selection. The results of this study provide new insights into the functional role of TP53 in the lactation of buffalo.
All relevant data are available within this paper and its Supplement.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-67-217-2024-supplement.
YM conceived and designed the research. LH and DS performed the material preparation and experiments. LH, DS, XF, and RG performed the data collection and analysis. LH and YM drafted the paper. All the authors read and approved of the final paper.
The contact author has declared that none of the authors has any competing interests.
Sample collection was implemented under the Guide for Animal Care and Use of Experimental Animals and was approved by the experimental animal management committee of Yunnan Agricultural University (no. YNAU2019llwyh019).
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research has been supported by the National Natural Science Foundation of China (grant nos. 32260822, 31460582, and 31760659) and the Natural Science Foundation Key Project of Yunnan Province, China (grant nos. 2014FA032 and 2007C0003Z).
This paper was edited by Henry Reyer and reviewed by two anonymous referees.
Abdel-Hamid, M., Huang, L., Huang, Z., Romeih, E., Yang, P., Zeng, Q., and Li, L.: Effect of Buffalo Breed on the Detailed Milk Composition in Guangxi, China, Foods, 12, 1603–1612, https://doi.org/10.3390/foods12081603, 2023.
Ahmad, S., Gaucher, I., Rousseau, F., Beaucher, E., Piot, M., Grongnet, J. F., and Gaucheron, F.: Effects of acidification on physico-chemical characteristics of buffalo milk: A comparison with cow's milk, Food Chem., 106, 11–17, https://doi.org/10.1016/j.foodchem.2007.04.021, 2008.
Ammah, A. A., Do, D. N., Bissonnette, N., Gévry, N., and Ibeagha-Awemu, E. M.: Co-expression network analysis identifies miRNA-mRNA networks potentially regulating milk traits and blood metabolites, Int. J. Mol. Sci., 19, 2500–2527, https://doi.org/10.3390/ijms19092500, 2018.
Appuhamy, J. A. D. R. N.: Regulatory roles of essential amino acids, energy, and insulin in mammary cell protein synthesis, PhD thesis, Virginia Tech, USA, 174 pp., 2010.
Bionaz, M. and Loor, J. J.: Gene networks driving bovine mammary protein synthesis during the lactation cycle, Bioinform. Biol. Insig., 5, 83–98, https://doi.org/10.4137/BBI.S7003, 2011.
Caron, A., Richard, D., and Laplante, M.: The roles of mTOR complexes in lipid metabolism, Annu. Rev. Nutr., 35, 321–348, https://doi.org/10.1146/annurev-nutr-071714-034355, 2015.
Carreño, D., Hervás, G., Toral, P. G., Castro-Carrera, T., and Frutos, P.: Fish oil-induced milk fat depression and associated downregulation of mammary lipogenic genes in dairy ewes, J. Dairy Sci., 99, 7971–7981, https://doi.org/10.3168/jds.2016-11019, 2016.
Chatchatee, P., Järvinen, K. M., Bardina, L., Vila, L., Beyer, K., and Sampson, H. A.: Identification of IgE and IgG binding epitopes on β-and κ-casein in cow's milk allergic patients, Clin. Exp. Allergy, 31, 1256–1262, https://doi.org/10.1046/j.1365-2222.2001.01167.x, 2001.
Fan, X., Qiu, L., Teng, X., Zhang, Y., and Miao, Y.: Effect of INSIG1 on the milk fat synthesis of buffalo mammary epithelial cells, J. Dairy Res., 87, 349–355, https://doi.org/10.1017/S0022029920000710, 2020.
Gang, X., Yang, Y., Zhong, J., Jiang, K., Pan, Y., Karnes, R. J., Zhang, J., Xu, W., Wang, G., and Huang, H.: P300 acetyltransferase regulates fatty acid synthase expression, lipid metabolism and prostate cancer growth, Oncotarget, 7, 15135–15149, https://doi.org/10.18632/oncotarget.7715, 2016.
Ghosh, A., Stewart, D., and Matlashewski, G.: Regulation of human p53 activity and cell localization by alternative splicing, Mol. Cell. Biol., 24, 7987–7997, https://doi.org/10.1128/MCB.24.18.7987-7997.2004, 2004.
Ghosh, H. S., McBurney, M., and Robbins, P. D.: SIRT1 negatively regulates the mammalian target of rapamycin, PLOS One, 5, 9199–9206, https://doi.org/10.1371/journal.pone.0009199, 2010.
Goldstein, I. and Rotter, V.: Regulation of lipid metabolism by p53-fighting two villains with one sword, Trends Endocrin. Met., 23, 567–575, https://doi.org/10.1016/j.tem.2012.06.007, 2012.
Hall, T. A.: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, Nucl. Acid. S., 41, 95–98, 1999.
Hallenborg, P., Feddersen, S., Francoz, S., Murano, I., Sundekilde, U., Petersen, R. K., Akimov, V., Olson, M. V., Lozano, G., Cinti, S., Gjertsen, B. T., Madsen, L., Marine, J. C., Blagoev, B., and Kristiansen, K.: Mdm2 controls CREB-dependent transactivation and initiation of adipocyte differentiation, Cell Death Differ., 19, 1381–1389, https://doi.org/10.1038/cdd.2012.15, 2012.
Hayashi, A. A., Nones, K., Roy, N. C., McNabb, W. C., Mackenzie, D. S., Pacheco, D., and McCoard, S.: Initiation and elongation steps of mRNA translation are involved in the increase in milk protein yield caused by growth hormone administration during lactation, J. Dairy Sci., 92, 1889–1899, https://doi.org/10.3168/jds.2008-1334, 2009.
Huang, J., Guesthier, M. A., and Burgos, S. A.: AMP-activated protein kinase controls lipid and lactose synthesis in bovine mammary epithelial cells, J. Dairy Sci., 103, 340–351, https://doi.org/10.3168/jds.2019-16343, 2020.
Horton, L. E., Bushell, M., Barth-Baus, D., Tilleray, V. J., Clemens, M. J., and Hensold, J.: p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation, Oncogene, 21, 5325–5334, https://doi.org/10.1038/sj.onc.1205662, 2002.
Jerry, D. J., Kuperwasser, C., Downing, S. R., Pinkas, J., He, C., Dickinson, E., Marconi S, and Naber, S. P.: Delayed involution of the mammary epithelium in BALB/c-p53null mice, Oncogene, 17, 2305–2312, https://doi.org/10.1038/sj.onc.1202157, 1998.
Kaelin, W. G.: The p53 gene family, Oncogene, 18, 7701–7705, https://doi.org/10.1038/sj.onc.1202955, 1999.
Kamada, R., Toguchi, Y., Nomura, T., Imagawa, T., and Sakaguchi, K.: Tetramer formation of tumor suppressor protein p53: structure, function, and applications, Biopolymers, 106, 598–612, https://doi.org/10.1002/bip.22772, 2016.
Khalil, H. S., Tummala, H., Chakarov, S., Zhelev, N., and Lane, D. P.: Targeting ATM pathway for therapeutic intervention in cancer, Biodiscovery, 1, 185–208, https://doi.org/10.7750/Biodiscovery.2012.1.3, 2012.
Kruse, J. P. and Gu, W.: SnapShot: p53 posttranslational modifications, Cell, 133, 930–930, https://doi.org/10.1016/j.cell.2008.05.020, 2008.
Kumar, S., Stecher, G., and Tamura, K.: MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 33, 1870–1874, https://doi.org/10.1093/molbev/msw054, 2016.
Lane, D. P.: p53, guardian of the genome, Nature, 362, 15–16, https://doi.org/10.1038/358015a0, 1992.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G.: Clustal W and Clustal X version 2.0, Bioinformatics, 23, 2947–2948, https://doi.org/10.1093/bioinformatics/btm404, 2007.
Leblanc, V. and May, P.: Activation and post-translational modifications of p53 following DNA damage, Med. Sci., 18, 577–584, https://doi.org/10.1051/medsci/2002185577, 2002.
Levine, A. J. and Oren, M.: The first 30 years of p53: growing ever more complex, 9, 49–758, Nat. Rev. Cancer, 9, 49–758, https://doi.org/10.1038/nrc2723, 2009.
Li, B., Greenberg, N., Stephens, L. C., Meyn, R., Medina, D., and Rosen, J. M.: Preferential overexpression of a 172 Arg→Leu mutant p53 in the mammary gland of transgenic mice results in altered lobuloalveolar development, Cell Growth Differ., 5, 711–721, 1994.
Li, H., Liu, X., Wang, Z., Lin, X., Yan, Z., Cao, Q., Zhao, M., and Shi, K.: MEN1/Menin regulates milk protein synthesis through mTOR signaling in mammary epithelial cells, Sci. Rep., 7, 5479–5487, https://doi.org/10.1038/s41598-017-06054-w, 2017.
Liu, L., Lin, Y., Liu, L., Wang, L., Bian, Y., Gao, X., and Li, Q.: Regulation of peroxisome proliferator-activated receptor gamma on milk fat synthesis in dairy cow mammary epithelial cells, In. Vitro. Cell. Dev.-An., 52, 1044–1059, https://doi.org/10.1007/s11626-016-0059-4, 2016.
Maddocks, O. D. and Vousden, K. H.: Metabolic regulation by p53, J. Mol. Med., 89, 237–245, https://doi.org/10.1007/s00109-011-0735-5, 2011.
Marijn, T. V., Jaarsveld, D., Deng, E. A., and Wiemer, Z. Z.: Tissue-specific chk1 activation determines apoptosis by regulating the balance of p53 and p21, Iscience, 12, 27–40, https://doi.org/10.1016/j.isci.2019.01.001, 2019.
Nasr, M., Awad, A., and El Araby, I. E.: Associations of leptin and pituitary-specific transcription factor genes' polymorphisms with reproduction and production traits in dairy buffalo, Reprod. Domest. Anim., 51, 597–603, https://doi.org/10.1111/rda.12726, 2016.
Nguyen, T. A., Menendez, D., Resnick, M. A., and Anderson, C. W.: Mutant TP53 posttranslational modifications: challenges and opportunities, Hum. Mutat., 35, 738–755, https://doi.org/10.1002/humu.22506, 2014.
Okita, N., Ishikawa, N., Mizunoe, Y., Oku, M., Nagai, W., Suzuki, Y., Matsushima, S., Mikami, K., Okado, H., Sasaki, T., and Higami, Y.: Inhibitory effect of p53 on mitochondrial content and function during adipogenesis, Biochem. Bioph. Res. Co., 446, 91–97, https://doi.org/10.1016/j.bbrc.2014.02.059, 2014.
Purushotham, A., Schug, T. T., Xu, Q., Surapureddi, S., Guo, X., and Li, X.: Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation, Cell Metab., 9, 327–338, https://doi.org/10.1016/j.cmet.2009.02.006, 2009.
Raj, N. and Attardi, L. D.: The transactivation domains of the p53 protein, CSH Perspect. Med., 7, 26047–26064, https://doi.org/10.1101/cshperspect.a026047, 2017.
Rius, A. G., Appuhamy, J., Cyriac, J., Kirovski, D., Becvar, O., Escobar, J., McGilliard, M. L., Bequette, B. J., Akers, R. M., and Hanigan, M., D.: Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids, J. Dairy Sci., 93, 3114–3127, https://doi.org/10.3168/jds.2009-2743, 2010.
Sambrock, J. and Russell, D.: Molecular cloning: A laboratory Manual, 3rd Edn., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001.
Saxton, R. A. and Sabatini, D. M.: mTOR signaling in growth, metabolism, and disease, Cell, 168, 960–976, https://doi.org/10.1016/j.cell.2017.02.004, 2017.
Schmid, P., Lorenz, A., Hameister, H., and Montenarh, M.: Expression of p53 during mouse embryogenesis, Dev. Brain Res., 113, 857–865, https://doi.org/10.1242/dev.113.3.857, 1991.
Shi, D. S., Wang, J., Yang, Y., Lu F. H., Li, X. P., and Liu, Q. Y.: DGAT1, GH, GHR, PRL and PRLR polymorphism in water buffalo (Bubalus bubalis), Reprod. Domest. Anim., 47, 328–334, https://doi.org/10.1111/j.1439-0531.2011.01876.x, 2012.
Singh, M., Aggarwal, S., Mohanty, A. K., and Mukhopadhyay, T.: Isolation, characterization and functional analysis of full length p53 cDNA from Bubalus bubalis, Gene, 568, 146–154, https://doi.org/10.1016/j.gene.2015.05.047, 2015.
Smith, T. M., Gilliland, K., Clawson, G. A., and Thiboutot, D.: IGF-1 induces SREBP-1 expression and lipogenesis in SEB-1 sebocytes via activation of the phosphoinositide 3-Kinase/Akt pathway, J. Invest. Dermatol., 128, 1286–1293, https://doi.org/10.1038/sj.jid.5701155, 2008.
Takasaki, M., Honma, T., Yanaka, M., Sato, K., Shinohara, N., Ito, J., Tanaka, Y., Tsuduki, T., and Ikeda, I.: Continuous intake of a high-fat diet beyond one generation promotes lipid accumulation in liver and white adipose tissue of female mice, J. Nutr. Biochem., 23, 640–645, https://doi.org/10.1016/j.jnutbio.2011.03.008, 2012.
Tilleray, V., Constantinou, C., and Clemens, M. J.: Regulation of protein synthesis by inducible wild-type p53 in human lung carcinoma cells, FEBS Lett., 580, 1766–1770, https://doi.org/10.1016/j.febslet.2006.02.030, 2006.
Vogelstein, B., Lane, D., and Levine, A. J.: Surfing the p53 network, Nature, 408, 307–310, https://doi.org/10.1038/35042675, 2000.
Wang, H. C., Ko, Y. H., Mersmann, H. J., Chen, C. L., and Ding, S. T.: The expression of genes related to adipocyte differentiation in pigs, J. Anim. Sci., 84, 1059–1066, https://doi.org/10.2527/2006.8451059x, 2006.
Wang, X. and Proud, C. G.: The mTOR pathway in the control of protein synthesis, Physiology, 21, 362–369, https://doi.org/10.1152/physiol.00024.2006, 2006.
Yahagi, N., Shimano, H., Matsuzaka, T., Najima, Y., Sekiya, M., Nakagawa, Y., Ide, T., Tomita, S., Okazaki, H., Tamura, Y., Iizuka, Y., Ohashi, K., Gotoda, T., Nagai, R., Kimura. S., Ishibashi, S., Osuga, J., and Yamada, N.: p53 Activation in adipocytes of obese mice, J. Biol. Chem., 278, 25395–25400, https://doi.org/10.1074/jbc.M302364200, 2003.
Yang, D. Q. and Kastan, M. B.: Participation of ATM in insulin signaling through phosphorylation of eIF-4E-binding protein 1, Nat. Cell Biol., 2, 893–898, https://doi.org/10.1038/35046542, 2000.
Yoshida, K., Liu, H., and Miki, Y.: Protein kinase C δ regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage, J. Biol. Chem., 281, 5734–5740, https://doi.org/10.1074/jbc.M512074200, 2006.
Zhang, X., Qin, Z., and Wang, J.: The role of p53 in cell metabolism, Acta Pharm. Sinic., 31, 1208–1212, https://doi.org/10.1038/aps.2010.151, 2010.