the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Bta-miR-33a affects gene expression and lipid levels in Chinese Holstein mammary epithelial cells
Ping Jiang
Ambreen Iqbal
Zhiqian Cui
Haibin Yu
Zhihui Zhao
MicroRNAs (miRNAs) are non-coding single-stranded RNA molecules of about 19–25 nucleotides in length that regulate different biological processes, including lipid metabolism. In this study, we explored the effect of bta-miR-33a on lipid metabolism in bovine mammary epithelial cells (BMECs) of Chinese Holstein for the first time. For this purpose, the plasmids of bta-miR-33a mimic, bta-miR-33a inhibitor and bta-miR-33a negative control were constructed to overexpress or repress bta-miR-33a in BMECs. The effects of plasmid transfection were analysed by examining the mRNA and protein expression levels of ELOVL6 and the intracellular triglycerides. The results showed that bta-miR-33a directly inhibited the expression of ELOVL6 in BMECs; decreased the mRNA levels of ELOVL5, HACD2, CPT1A and MSMO1; and increased the mRNA level of ALOX15. Sequence bta-miR-33a also increased the contents of triglycerides in the cells, presumably as a consequence of these gene expression changes. In summary, the results of the present study suggest that bta-miR-33a regulates lipid metabolism by targeting ELOVL6, which might be a potential molecular marker of milk fat composition.
- Article
(1921 KB) - Full-text XML
- BibTeX
- EndNote
Milk, a white liquid food that is highly rich in fat, protein, vitamins and other nutrients, is produced in mammary glands to serve as a primary source of nutrition for mammalian progeny (Winckel et al., 2011). Due to its nutrient-rich composition, it is also consumed by adult humans to meet their dietary requirements. In terms of commercial production, cow milk contributes the largest amount, by volume, with respect to milk production worldwide (Gerosa and Skoet, 2012).
MicroRNAs (miRNAs) are endogenous non-coding RNA molecules of 19–25 nucleotides in length that are highly conserved and can form complementary or incomplete complementary binding to the 3′UTR of target messenger RNAs (mRNAs) to control gene expression via mRNA degradation or translational inhibition. As such, miRNAs play essential roles in many physiological or pathological processes, such as cell metabolism, stress responses and fat metabolism (Bartel, 2004; Ambros, 2004). Previous research has reported that many miRNAs have roles in lipid and glucose metabolism, including miR-27 (Lin et al., 2013), miR-33 (Dávalos et al., 2011), miR-370 (Iliopoulos et al., 2010), miR-378/378* (Gerin et al., 2010) and miR-2885 (Benatti et al., 2014). To further explore the regulatory roles of miRNAs in milk fat synthesis, our laboratory has previously performed differential miRNA expression analysis between bovine mammary epithelial cells (BMECs) from high-milk-fat and low-milk-fat Chinese Holstein cows. Among all of the differentially expressed miRNAs, bta-miR-33a was found to have low expression in the BMECs of low-milk-fat cows and high expression in the BMECs of high-milk-fat Chinese Holstein cows (Shen et al., 2016). One of the target genes of bta-miR-33-a is the elongation of very long chain fatty acid, like fatty acid elongase 6 (ELOVL6), a member of the conserved endoplasmic-reticulum-localised enzyme family involved in the elongation process of long-chain fatty acids (Leonard et al., 2004). The ELOVL family member 6 (ELOVL6) is highly expressed in white adipose tissue, brown adipose tissue, skin, testis, brain, liver and adrenal gland, and it is also involved in the regulation of steroidogenesis and lipogenesis (Moon et al., 2001). The high expression of ELOVL6 in caprine and bovine mammary tissues highlights its role in milk fat metabolism (Shi et al., 2015). Further, dietary supplementation of cows with rapeseed and sunflower oil was found to alter the expression of ELOVL6 in mammary tissue, suggesting its lipid metabolism regulatory role in the mammary glands (Leroux et al., 2016). Recently, ELOVL6 was found to play a role in the elongation of long-chain saturated fatty acids in caprine mammary epithelial cells (Shi et al., 2017). Further, it was also found to affect the fatty acid composition in bovine adipocytes (Junjvlieke et al., 2020).
The family of miR-33 consists of two members, including miR-33a and miR-33b; both have the same seed sequence except for the two bases (Lu et al., 2018). The location of miR-33a is in intron 15 of sterol regulatory element-binding transcription factor 2 (SREBF2), and miR-33b is located in intron 17 of sterol regulatory element-binding transcription factor 1 (SREBF1). Both play a vital role in lipid hemostasis by regulating the expression of their target genes (Horie et al., 2014). Previous research has proven that miR-33a, through its target genes, plays a critical role in regulating cholesterol efflux, insulin signalling, fatty acid oxidation and high-density lipoprotein (HDL) formation. miR-33a inhibits cholesterol transport in macrophages by targeting the 3′UTR of the mRNA of the ATP binding cassette subfamily A member 1 (ABCA1) gene (Niu et al., 2018). miR-33a modulates intracellular fatty acid oxidation in humans by targeting the mRNA of the hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta (HADHB) gene (Rayner et al., 2012). The miR-33a target genes carnitine palmitoyl transferase 1A (CPT1A) and HADHB enhance the mitochondrial oxidative capacity and ATP production as well as increasing plasma HDL. Further, CPT1A and HADHB also reduce very low density lipoprotein receptor (VLDL) and triglyceride levels, providing a promising therapeutic strategy for treating dyslipidemia, which increases the risk of cardiovascular diseases (Rottiers et al., 2013). The treatment of anti-miR-33 in monkeys has been shown to result in a 4-fold increase in the mRNA expression of protein kinase, AMP-activated, alpha 1 (PRKAA1) in the liver tissue (Rayner et al., 2011). Notably, miR-33a also targets insulin receptor substrates that are important components of the insulin-signalling pathway in the liver; thus, overexpression of miR-33a in a hepatic cell line reduced insulin receptor substrate 2 (IRS2) levels and inhibited the activation of its downstream messenger cascade (Dávalos et al., 2011). Therefore, miR-33a may regulate the AMP-activated protein kinase (AMPK) pathway, cholesterol transport, fatty acid oxidation and insulin-signalling pathways, suggesting that miR-33a plays a crucial role in regulating lipid metabolism (Dávalos et al., 2011).
Despite much research on miRNAs and their role in milk fat metabolism in dairy cattle, bta-miR-33a and its target genes have not been studied to date. Thus, this study focused on miR-33a in order to better understand the involvement of bta-miR-33a in the milk fat metabolism of Chinese Holstein cows. Through bioinformatic analysis, we predicted ELOVL6 as a target gene of bta-miR-33a. By altering the expression of bta-miR-33a in BMECs, ELOVL6 and the functions of bta-miR-33a in triglyceride metabolism were examined. The results revealed the regulatory roles and molecular mechanisms between bta-miR-33a and ELOVL6 in milk fat synthesis.
2.1 Cell source
BMECs from Chinese Holstein dairy cows were maintained at the Animal Genetics and Breeding Laboratory, College of Animal Science, Jilin University (Lu et al., 2012). Chinese Holstein cows with a low-milk-fat content were utilised to obtain the BMECs used in this investigation. The mammary gland tissue from freshly slaughtered cows was put into a 5 % () penicillin–streptomycin phosphate-buffered saline (PBS) solution (Shanghai Yuanye Bio-Technology Co., Ltd.), and the temperature was maintained at 37 ∘C following immediate transfer of the sample to the laboratory. The tissue was washed many times with PBS, in order to remove debris and lipids, until the tissue solution was no longer turbid. The tissue was cut into 1 mm3 pieces and washed with PBS. A tiny bit of tissue was cut and put into a Petri dish. The basal medium of Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12, 1:1; HyClone, Logan, USA), which comprised 10 % () fetal bovine serum (FBS; Tian hang, Zhejiang, China), 1 % penicillin–streptomycin (Shanghai Yuanye Bio-Technology Co., Ltd.) and 1 % () epithelial growth factor (Thermo Fisher Scientific Shanghai Instruments, China), was added to boost cell proliferation. Between one and three drops of basal media were applied to the surrounding tissue to protect the cells from drying out, and the Petri dish was placed in an incubator at 37 ∘C with 5 % CO2 for 6 h. After 6 h, 5–6 mL of basal medium was added, ensuring that the cells remained on the substrate and did not float on the upper part of the Petri dish. After 48 h, the medium was replaced again until the plate was confluent with cells. Finally, the cells were harvested, using 0.02 % ethylenediaminetetraacetic acid (EDTA) and 0.25 % trypsin, and placed onto a fresh culture plate in order to eliminate the other cell types. After three to five passages, the pure mammary gland cells were attained (Lu et al., 2012). All of the experiments were performed according to the guidelines for the care and use of laboratory animals of Guangdong Ocean University.
2.2 Bioinformatic prediction of the bta-miR-33a target gene
An in silico analysis was performed to procure the mature sequence of bta-miR-33a using miRBase (http://www.mirbase.org, last access: 12 September 2021). TargetScan (http://www.targetscan.org, last access: 12 September 2021) was consulted to predict its target genes with binding sites for miR-33a and to obtain the 3′UTR sequence of target genes from the GenBank database (https://www.ncbi.nlm.nih.gov/, last access: 12 September 2021). The mining of functional attributes of ELOVL6 protein was accomplished using STRING version 11.0 (https://string-db.org/, last access: 12 September 2021). The primers for target genes were designed using Primer Premier 6.0 (PREMIER Biosoft International, Canada). The bta-miR-33a mimic (GTGCATTGTAGTTGCATTGCA), bta-miR-33a inhibitor (TGCAATGCAACTACAATGCAC) and miRNA-shNC (AAATGTACTGCGCGTGGAGAC) used were synthesised by Shanghai GenePharma Co., Ltd. The plasmids were verified by sequencing (Fig. A1). Each experimental step/reaction was repeated three times using β-actin and U6 as the reference genes. Primers were designed according to the 3′UTR sequence information (Table 1).
Table 1Primer sequences of bta-miR-33a and candidate target genes for qPCR.
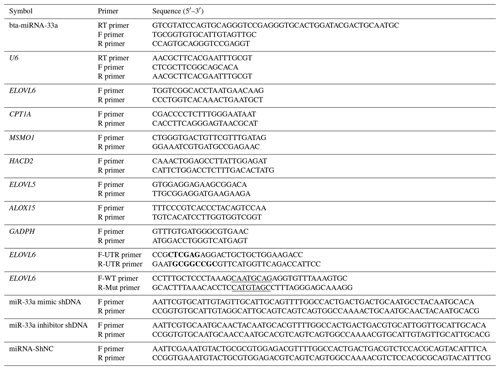
Bold font represents the enzyme cleavage site, and underlined text represents an miRNA binding sequence or mutation sequence. The abbreviations used in the primer column are as follows: F primer – forward primer, R primer – reverse primer, and RT primer – reversed transcript primer.
2.3 Cell culture and transfection reagents
BMECs were cultured in a basic culture medium containing DMEM/F12 (1:1) (HyClone, Logan, USA), 10 % fetal bovine serum (Tian hang, Zhejiang, China) and 1 % penicillin–streptomycin (Shanghai Yuanye Bio-Technology Co., Ltd.) in cell culture dishes at 37∘ in an atmosphere of 5 % CO2. BMECs were seeded into six-well cell culture plates (Corning, USA) with 1×106 cells per well for transfection. When the confluency of BMECs reached approximately 70 %, the cell culture medium was changed to a fresh growth medium without antibiotics. Briefly, the transfection mixture of 150 µL of Opti-MEM serum-free media, containing 3 µg of the bta-miR-33a-mimic, negative-control or bta-miR-33a-inhibitor plasmids as well as 7 µL of FuGENE HD transfection reagent (Promega Biotechnology Co., Ltd., Beijing), was incubated at room temperature for 15 min according to the instructions provided by the supplier, transferred into six-well cell culture plates with BMECs, and incubated at 37∘ in an atmosphere of 5 % CO2 for 24 h. The transfection efficiency after 24 h was evaluated by green fluorescent protein (GFP) expression under a fluorescence microscope (TE2000, Nikon, Tokyo, Japan). Three replicates of the experiments were carried out using the identical plasmids by transfecting equal numbers of cells in the various wells. The bta-miR-33a-mimic, negative-control and bta-miR-33a-inhibitor plasmids were constructed by GenePharma (Suzhou, China).
2.4 Extraction of RNA and qRT-PCR
The cells transfected with the bta-miR-33a-mimic, negative-control or bta-miR-33a-inhibitor plasmids were harvested 24 h post-transfection. Total RNA was then extracted using TRIzol reagent (catalogue no. 15596-026, Invitrogen, USA), following the manufacturer's protocol. The integrity and purity of the RNA were confirmed using 1.5 % agarose gel electrophoresis (80 V for 15 min) and a spectrophotometer (optical density, OD, values from 1.8 to 2.2) respectively. Then, 1 µg of total RNA was reverse transcribed to cDNA using the PrimeScript™ RT reagent kit with genomic DNA (gDNA) eraser (Takara Biological Company, Japan). The qRT-PCR was performed using Mx3005P (Agilent Stratagene, USA) in a 20 µL reaction volume, including 10.0 µL of 2× SYBR® Premix Ex Taq (Tli RNaseH Plus, Takara Bio Inc., Dalian, China), 0.5 µL of forward and reverse primers (10 µM), 2.0 µL of template cDNA, and 7 µL of nuclease-free water. The real-time PCR conditions were 95 ∘C for 5 min, 40 cycles of 95 ∘C for 10 s and 59 ∘C for 30 s. The relative expression levels were calculated using the 2−ΔΔCt cycle. GADPH was used as the internal reference gene in the experiments, as our previous studies have shown that GADPH is stably expressed in bovine mammary epithelial cells (Iqbal et al., 2022). Each experimental step/reaction was repeated three times.
2.5 Luciferase reporter assay
The bta-miR-33a sequence was cloned into the pmiR-RB-REPORT™ luciferase reporter plasmid using restriction enzymes XhoI and NotI (New England Biolabs, Beijing, China), and the plasmid was named “pmiR-RB-REPORT™-ELOVL6-3′UTR-WT”. To construct the mutated reporter plasmid, the bta-miR-33a putative seed binding site 5′CAATGCAG3′ in pmiR-RB-REPORT™-ELOVL6-3′UTR-WT was mutated to the sequence 5′CATGTAGC 3′ by site-directed mutagenesis, which resulted in the plasmid pmiR-RB-REPORT™-ELOVL6-3′UTR-MUT. The target fragment connected to the pmiR-RB-REPORT™ vector was transformed into competent cells to obtain a recombinant plasmid. After double-digestion and sequencing identification, luciferase activity was detected using a SpectraMax M5 microplate reader (Molecular Devices, USA). Normalised relative light units (RLUs) were determined using a dual-luciferase reporter assay system (Promega Corp, USA), following the manufacturer's instructions, and the relative luciferase activity of the firefly (hluc+) and Renilla (hRluc) luciferase were detected. The hluc+ luciferase was used as the reference to correct for variations in transfection efficiency, and the relative luciferase activity was calculated with the following equation: relative luciferase activity = hRluc hluc+. Three replicates of the experiments were carried out by transfecting equal numbers of cells using the same vectors in various wells.
2.6 Western blot analysis
The bta-miR-33a mimic, bta-miR-33a inhibitor and bta-miR-33a negative control were transfected into BMECs, and the effect of the transfections on protein expression was analysed. Total protein was extracted at 24 h post-transfection using radioimmunoprecipitation assay (RIPA) buffer (Boster, Wuhan, China), and the supernatant was collected in a centrifuge tube. Protein concentration was determined with a bicinchoninic acid (BCA) protein quantitation assay (Nanjing KeyGen Biotech. Co. Ltd.) using a spectrophotometer (UNIC2802H, Shanghai, China), according to the given protocol. For western blotting, equal amounts of protein were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories, USA). Immunoblotting was performed with primary polyclonal ELOVL6 antibody (ab69857, Abcam, USA) and monoclonal anti-β-actin (β-actin, ab8227, Abcam, USA) at a 1:500 dilution and a 1:1000 dilution respectively. The horseradish peroxidase (HRP) conjugated anti-rabbit secondary antibody was used at a 1:2000 dilution (BS13271, BioWorld, USA). The immunoblots were developed using an ECL advance western blotting detection kit (Thermo, USA), and the signal intensities were captured by a Tanon 5200 chemiluminescence/fluorescence image analysis system (Tanon, China).
2.7 Detection of the intracellular triglyceride level
For triglyceride extraction, the transfection of plasmids of miR-33a mimic, inhibitor and negative control was carried out in BMECs in six-well cell culture plates. The intracellular triglyceride level of BMECs was extracted and detected using a tissue and cell triglyceride assay kit (APPLYGEN, Beijing, China), following the manufacturer's protocols. A total of 10 µL of sample was used to estimate the protein concentration. The sample was diluted 10 times, and protein (BCA, protein quantitation assay, Nanjing KeyGen Biotech. Co. Ltd.) was measured at 562 nm using Gen5 CHS software according to the protocol. The quantity of total protein was used to adjust the triglyceride level. Each sample was repeated three times, and the average values were used to calculate the final results.
2.8 Statistical analysis
The results were analysed using unpaired t tests between groups in GraphPad Prism 6 software (San Diego, CA, USA). All results are presented as the means ± standard error of the mean (SEM) of separate experiments (N≥3). Statistical significance is presented as * p<0.05, p<0.01 and p<0.001.
3.1 Expression of bta-miR-33a in BMECs
After 24 h of transfection of plasmids of bta-miR-33a mimic, bta-miR-33a inhibitor and negative control into BMECs, the BMECs were observed under a fluorescent microscope to determine the transfection efficiency (Fig. 1a). The results indicated transfection success, and the transfection efficiency was up to 50 %. The relative expression level of bta-miR-33a was analysed in bta-miR-33a mimic cells, bta-miR-33a inhibitor cells, and bta-miR-33a negative-control cells using qRT-PCR (Fig. 1b). The results showed that the relative expression of miR-33a in bta-miR-33a-mimic-transfected BMECs was significantly higher compared with that of the negative-control cells (Fig. 1b), whereas the relative expression of bta-miR-33a-inhibitor-transfected BMECs was significantly lower compared with that of the negative-control cells (Fig. 1b). These bta-miR-33a relative expression results of transfected BMECs in three experimental groups confirmed the successful transfection of plasmids of the bta-miR-33a mimic, bta-miR-33a inhibitor and bta-miR-33a negative control.
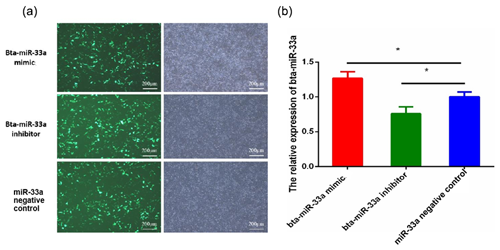
Figure 1Transfection of bovine mammary epithelial cells (BMECs) with plasmids of the bta-miR-33a mimic, bta-miR-33a inhibitor and bta-miR-33a negative control: (a) microscopy of cultured BMECs showing transfection efficiency (left panels) and cell morphology (right panels); (b) post-transfection relative expression of bta-miR-33a in the three different experimental groups of BMECs. Experimental data are shown as mean ± SEM. “*” represents p<0.05 (N=3).
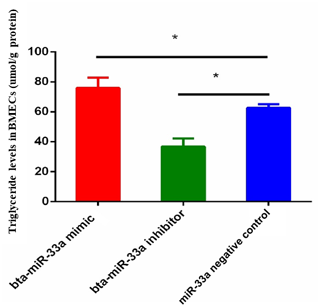
3.2 Effect of bta-miR-33a on triglyceride level in BMECs
The triglyceride metabolism in BMECs of Chinese Holstein cattle directly affects the milk fat composition. The analysis of triglyceride levels in the transfected BMECs showed that the triglyceride level in BMECs of the bta-miR-33a-mimic group was significantly higher than the cells in the negative-control group. In comparison, significantly lower levels of triglycerides were observed in the bta-miR-33a-inhibitor-transfected BMECs compared with the negative-control cells (Fig. 2). These results strongly suggested that bta-miR-33a positively regulated triglyceride synthesis in the BMECs.
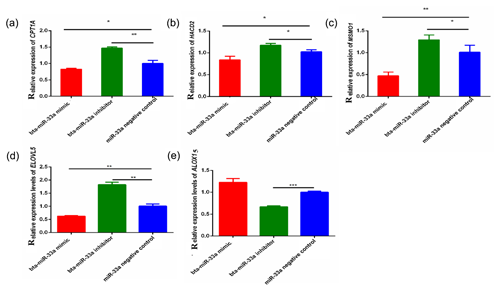
3.3 Expression of bta-miR-33a target genes related to lipid metabolism
Through bioinformatical prediction, more than a dozen target genes related to lipid metabolism were screened in addition to ELOVL6. From these target genes predicted in silico, we selected five candidate target genes that have a role in milk fat metabolism in the BMECs of Chinese Holstein cattle. The relative expression of these selected genes showed that the expression of the CPT1A, ELOVL5, MSMO1 and HACD2 genes was significantly down-regulated in the bta-miR-33a-mimic-transfected BMECs compared with the negative-control BMECs (Fig. 3a, b, c, d), whereas ALOX15 gene expression was significantly up-regulated (Fig. 3e). In addition, the expression of CPT1A, ELOVL5, MSMO1 and HACD2 was up-regulated in the bta-miR-33a-inhibitor-transfected BMECs compared with the negative-control BMECs (Fig. 3a, b, c, d), whereas ALOX15 gene expression was down-regulated (Fig. 3e). Thus, overall, bta-miR-33a was shown to positively regulate the gene expression of ALOX15 and negatively regulate the gene expression of CPT1A, ELOVL5, MSMO1 and HACD2.
3.4 Sequence bta-miR-33a regulates the mRNA expression of ELOVL6 by specifically binding to its 3′UTR
The mature sequence of bta-miR-33a was identified from miRBase (http://www.mirbase.org, last access: 12 September 2021), and the binding sites for bta-miR-33a in 3′UTR of ELOVL6 were found using TargetScan (http://www.targetscan.org, last access: 12 September 2021) (Fig. 4a). Amplified fragments that included the 3′UTR target sites (Fig. 4b) were double-digested with restriction enzymes (Fig. 4c) and sequenced (Fig. 4e) to identify the construction of wild-type and mutant-type vectors based the original vector backbone of pmiR-RB-REPORT™ (Fig. 4d). A luciferase reporter assay was used to verify the bta-miR-33a binding site in the 3′UTR of ELOVL6 mRNA. The bta-miR-33a-mimic plasmid with pmiR-RB-REPORT™-ELOVL6-3′UTR-WT and pmiR-RB-REPORT™-ELOVL6-3′UTR-MUT plasmids were transiently co-transfected into BMECs respectively. The relative luciferase activity suggests that the luciferase activity of the pmiR-RB-REPORT™-ELOVL6-3′UTR-WT group was significantly decreased (p<0.01) compared with the pmiR-RB-REPORT™-ELOVL6-3′UTR-MUT and the negative-control cells. The luciferase activity in pmiR-RB-REPORT™-ELOVL6-3′UTR-MUT cells was significantly increased (p<0.01) compared with the pmiR-RB-REPORT™-ELOVL6-3′UTR-WT cells (Fig. 4f). The combined bioinformatic and luciferase activity results suggest that bta-miR-33a specifically targets the 3′UTR sequence of ELOVL6 mRNA, although some off-target activity was seen in the sequence of mutated 3′UTR.
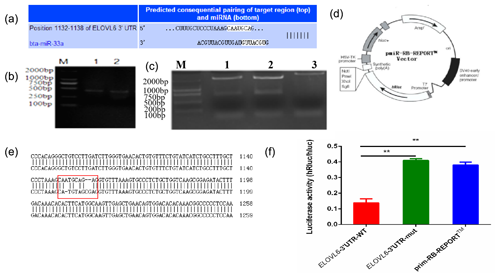
Figure 4Validation of bta-miR-33a and its predicted target gene ELOVL6 via (a) bioinformatic prediction; (b) electrophoresis of PCR amplification fragments, including the 3′UTR target site; (c) double-restriction enzyme digestion gel electrophoresis, lane 1 ELOVL6 wild-type vector, lane 2 ELOVL6 mutant vector and lane 3 pmiR-RB-REPORT™ vector; (d) original vector backbone of pmiR-RB-REPORT™, using the restriction sites XhoI and NotI respectively; (e) sequence alignment of ELOVL6 wild-type and mutant dual-luciferase reporter gene vectors; and (f) luciferase activity reporter assay. The experimental data are shown as the mean ± SEM. “” represents p<0.01 (N=3).
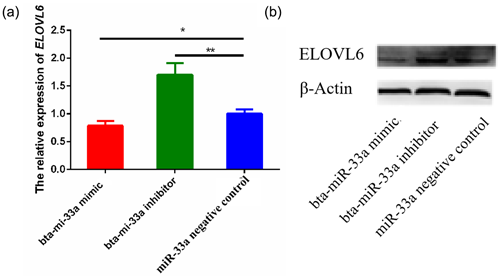
3.5 Effects of bta-miR-33a on mRNA and protein levels of ELOVL6
After verifying ELOVL6 as a target gene of bta-miR-33a in the BMECs of Chinese Holstein cows, we investigated the mRNA and protein expression of ELOVL6 in BMECs transfected with bta-miR-33a-mimic, bta-miR-33a-inhibitor and negative-control plasmids. The qRT-PCR results showed that the mRNA expression of ELOVL6 was significantly down-regulated (p<0.05) in BMECs of the bta-miR-33a-mimic-transfected group compared with the bta-miR-33a-inhibitor and negative-control cells (Fig. 5a). Moreover, the expression of ELOVL6 was significantly up-regulated in BMECs of the bta-miR-33a-inhibitor-transfected group compared with the bta-miR-33a-mimic and negative-control cells (p<0.05). The comparison of bta-miR-33a-inhibitor and negative-control cells showed that the expression ELOVL6 was significantly higher in bta-miR-33a-inhibitor BMECs compared with the negative-control BMECs (p<0.05, Fig. 5a). To further verify the effect of transfection of bta-miR-33a mimic and bta-miR-33a inhibitor on the protein levels of ELOVL6, western blotting was conducted (Fig. 5b). The results were similar to the mRNA levels of ELOVL6 for the three experimental groups: the level of ELOVL6 protein was higher in the BMECs transfected with the bta-miR-33a-inhibitor group compared with the mimic and negative-control cells (Fig. 5b). In addition to the luciferase reporter assay, these results further verify ELOVL6 as a target gene of bta-miR-33a and reveal that bta-miR-33a negatively regulates ELOVL6 expression.
3.6 STRING interaction network of ELOVL6 in Bos taurus
The STRING interaction network (https://string-db.org/cgi/network.pl?taskId=JhI2SG3gdQo7, last access: 12 September 2021; https://cn.string-db.org/cgi/network?taskId=bXbGtelvjFpC&sessionId=bavFa9mdqMHo, last access: 12 September 2021) shows that the ELOVL6 protein is involved in many functions related to lipid metabolism, including the fatty acid biosynthesis process, acyl-CoA biosynthesis, acetyl-CoA metabolism and other related processes. The main enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of ELOVL6 include fatty acid metabolism, biosynthesis of unsaturated fatty acids, fatty acid elongation, fatty acid biosynthesis and the AMPK signalling pathway. These functions are carried out through the interaction of ELOVL6 with other genes involved in lipid metabolism (Fig. 6). A comparison of the ELOVL6 string interaction in Bos taurus and Homo sapiens showed that the function of some genes, including HSD17B12, SREBF1A and HACD2(PTPLB), in the regulation of lipid metabolism is conserved across these two species. Thus, the overexpression and repression of bta-miR-33a in BMECs may drive lipid metabolism directly by regulating ELOVL6 expression or indirectly via the interlinked functions of other genes related to the lipid metabolism.
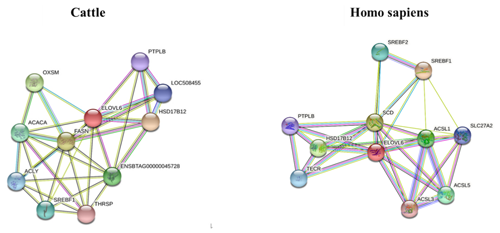
Figure 6The STRING interaction network of ELOVL6 in Bos Taurus (https://string-db.org/cgi/network.pl?taskId=JhI2SG3gdQo7, last access: 12 September 2021) and Homo sapiens (https://cn.string-db.org/cgi/network?taskId=bXbGtelvjFpC&sessionId=bavFa9mdqMHo, last access: 12 September 2021), showing 3-oxoacyl-ACP synthase, mitochondrial (OXSM); 3-hydroxyacyl-CoA dehydratase 2 (HACD2, PTPLB); acetyl-CoA carboxylase alpha (ACACA); acyl-CoA synthetase long-chain family member 1 (ACSL1); acyl-CoA synthetase long-chain family member 3 (ACSL3); acyl-CoA synthetase long-chain family member 5 (ACSL5); ATP citrate lyase (ACLY); ELOVL fatty acid elongase 6 (ELOVL6); fatty acid synthase (FASN); hydroxysteroid 17-beta dehydrogenase 12 (HSD17B12); sterol regulatory element-binding transcription factor 1 (SREBF1); sterol regulatory element-binding transcription factor 2 (SREBF2); stearoyl-CoA desaturase (SCD); solute carrier family 27, member 2 (SLC27A2); thyroid hormone responsive (THRSP); and trans-2,3-enoyl-CoA reductase (TECR).
miRNAs play significant roles in the growth and development of mammary glands and milk production in dairy cattle (Shen et al., 2016). The miRNAs work together in the same and different biological pathways to affect the target genes (Xu et al., 2016). Regarding their function in lipid metabolism, many miRNAs have been found to play essential roles. For example, miR-22 can regulate fatty acid metabolism by influencing cholesterol synthesis and fatty acid β-oxidation (Esau et al., 2006), whereas miR-370 down-regulates the expression of CPT1A, leading to reduced fatty acid β-oxidation (Iliopoulos et al., 2010). Previous work on lipid metabolism in BMECs has shown that bta-miR-124a can also affect triglycerides and free fatty acids by regulating the peroxisomal trans-2-enoyl-CoA reductase (PECR) gene (Shen et al., 2019). miR-21-3p can affect the formation of triglycerides in BMECs by targeting the ELOVL5 gene (Li et al., 2019). The previous work of our laboratory reported that miR-152 could significantly influence triglyceride production by targeting the acetyl-CoA acyltransferase 2 (ACAA2) and hydroxysteroid 17-beta dehydrogenase 12 (HSD17B12) genes (Yang et al., 2018). miR-29 also influences triglyceride production by targeting the lipoprotein lipase (LPL) and thymine DNA glycosylase (TDG) genes, resulting in mammary epithelial cell apoptosis and triglyceride synthesis (Yang et al., 2016).
Our group has already performed miRNA sequencing on the mammary epithelial cells of two Chinese Holstein cows: one with a high milk fat percentage and another with low milk fat percentage. Among differentially expressed miRNAs, bta-miR-33a was also found, hinting at its role in regulating milk fat metabolism. Bioinformatical analysis showed that the ELOVL6 gene contains a binding site for bta-miR-33a in its 3′UTR, which was verified in a luciferase reporter assay. To further explore the effects of bta-miR-33a on lipid metabolism in BMECs, the BMECs were transfected with bta-miR-33a-mimic, bta-miR-33a-inhibitor and negative-control plasmids. The results show that bta-miR-33a negatively regulates the expression of ELOVL6.
Additionally, bta-miR-33a positively regulates the expression of ALOX15, whereas it negatively regulates the expression of HACD2, CPT1A, ELOVL5 and MSMO1. The effect of bta-miR-33a transfection on triglyceride levels in three experimental cell groups indicated that the triglyceride level was higher in the bta-miR-33a-mimic cells than in the inhibitor and negative-control cells. In contrast, the triglyceride level in the inhibitor cells was low compared with the mimic and negative-control cells. These results delineated the role of bta-miR-33a in regulating the expression of ELOVL6 and other genes related to milk fat metabolism, ultimately affecting the triglyceride production in BMECs.
ELOVL6 belongs to the elongation of very long chain fatty acid gene family that encode enzymes to synthesise very long chain fatty acids in mammals. The ELOVL6 protein controls the rate-limiting step in long-chain fatty acid synthesis, which promotes the extension of C12, C14 and C16 fatty acids (Bond et al., 2016) as well as being associated with various pathophysiological functions related to fat metabolism, including insulin resistance, atherosclerosis and non-alcoholic steatohepatitis (Matsuzaka et al., 2007; Muir et al., 2013). In ELOVL6 mice, the liver accumulates significantly more triglycerides than in wild-type mice when fed a fat-free/high-carbohydrate diet, suggesting that the deletion of ELOVL6 can increase the levels of triglycerides in the liver (Matsuzaka et al., 2007; Moon et al., 2015). This is consistent with the result in bta-miR-33a-mimic BMECs: the down-regulation of ELOVL6 increased the level of triglycerides in the BMECs. However, knockdown of ELOVL6 negatively affected triglyceride level in goat mammary epithelial cells (Shi et al., 2017). This is inconsistent with the results of bta-miR-33a overexpression herein, which showed increased intracellular triglycerides through the down-regulation of ELOVL6 expression levels by directly targeting its 3′ untranslated region. Thus, we speculate that bta-miR-33a also alters the expression of other genes related to fat metabolism to increase the intracellular triglycerides.
The results of this work suggests that overexpression of bta-miR-33a also up-regulates the expression of ALOX15 and down-regulates ELOVL5, HACD2, CPT1A and MSMO1, consistent with our previous study in BMECs from high-milk-fat and low-milk-fat Chinese Holstein cows (Shen et al., 2016). ELOVL5, another member of the ELOVL family of enzymes, regulates the elongation of polyunsaturated C18 and C20 fatty acids (Green et al., 2010). Recently, ELOVL5 was found to regulate the synthesis of long-chain unsaturated fatty acids in goat mammary epithelial cells (Shi et al., 2017). The down-regulation of ELOVL5 and ELOVL6 in the bta-miR-33a-mimic BMECs increased the triglyceride levels in these cells in this study and may indicate the involvement of ELOVL5 in the regulation of triglyceride level in addition to its role in fatty acid elongation.
The CPT1A protein is located in the mitochondrial outer membrane and transports short-chain and long-chain fatty acids into the mitochondria for β-oxidation (Lee et al., 2011). This transport is the rate-limiting step for fatty acid β-oxidation and directly affects triglyceride levels (Lee et al., 2011). A recent study showed that bta-miR-33a down-regulates target genes, resulting in the enhancement of the mitochondrial oxidative capacity and ATP production in humans, which may activate triglyceride and fatty acids hydrolysis, thereby decreasing triglyceride levels in cells (Kwon et al., 2016). This is consistent with the results of this work, which demonstrated that CPT1A is a target gene of bta-miR-33a (Dávalos et al., 2011). Interestingly, CPT1A has been reported as a target gene of bta-miR-33a in humans as well. However, no other reports about the target relationship between bta-miR-33a and CPT1A in cattle exist, so this needs further elucidation. ALOX15 mainly regulates lipid metabolism by being involved in the synthesis of polyunsaturated fatty acids; it has been also found that ALOX15 is required for efferocytosis of apoptotic adipocytes by macrophages in vitro (Kwon et al., 2016). MSMO1 is another important gene in fatty acid biosynthesis, and also functions in cholesterol biosynthesis (He et al., 2011, 2014). These three genes were down-regulated by bta-miR-33a and are all involved in the synthesis and oxidation fatty acids. Therefore, the alteration in triglyceride level in the different BMEC groups may be directly regulated by bta-miR-33a or indirectly regulated via the regulation of other genes or pathways; this requires further validation.
Sequence bta-miR-33a binds to a specific sequence on 3′UTR of ELOVL6 and directly inhibits the expression of ELOVL6 in Chinese Holstein mammary epithelial cells. Sequence bta-miR-33a also affected the expression of five other genes related to fat metabolism, namely ELOVL5, HACD2, CPT1A, MSMO1 and ALOX15. Lastly, bta-miR-33a promoted the production of triglycerides in these cells, most likely as a result of these effects on gene expression. The function of bta-miR-33a has a specific effect on milk fat levels in the mammary epithelial cells of dairy cows, providing a promising strategy for improving milk quality.
Figure A1GenePharma miRNA expression vector report: (a) sequencing results of miR-33a mimic; (b) sequencing results of miR-33a inhibitor; (c) sequencing results of miRNA negative-control plasmid; (d) the miRNA expression empty vector (backbone vector) used for constructing expression plasmids by the GenePharma company; (e) sequencing peak map of miR-33a mimic; and (f) the sequencing peak map of miR-33a inhibitor. The bta-miR-33a mimic, bta-miR-33a inhibitor and miRNA-shNC were synthesised by Shanghai GenePharma Co., Ltd. The plasmids were verified though sequencing by Sangon Biotech Shanghai Co. Ltd.
No data sets were used in this article.
HY was responsible for conceptualising the study and carrying out the formal analysis. ZZ acquired funding and was responsible for project administration. PJ developed the methodology. ZC acquired resources. PJ and AI wrote the original draft of the paper and were also responsible for reviewing and editing the paper. All authors have read and agreed upon the published version of the paper.
The contact author has declared that none of the authors has any competing interests.
All of the experiments were performed according to the guidelines for the care and use of laboratory animals of Guangdong Ocean University.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was supported by the National Natural Science Foundation of China (grant nos. 32002165 and 32072717), the Innovative Strong School Engineering Youth Talent Project of the Department of Education of Guangdong Province (grant no. 2019KQNCX042) and the Innovative Strong School Engineering Key Platform Project of the Department of Education of Guangdong Province (grant no. 2018302).
This research has been supported by the National Natural Science Foundation of China (grant nos. 32002165 and 32072717) and the Department of Education of Guangdong Province (grant nos. 2019KQNCX042 and 2018302).
This paper was edited by Antke-Elsabe Freifrau von Tiele-Winckler and reviewed by two anonymous referees.
Ambros, V.: The functions of animal microRNAs, Nature, 431, 350–355, https://doi.org/10.1038/nature02871, 2004.
Bartel, D. P.: MicroRNAs: Genomics, Biogenesis, Mechanism, and Function, Cell, 116, 281–297, https://doi.org/10.1016/S0092-8674(04)00045-5, 2004.
Benatti, R. O., Melo, A. M., Borges, F. O., Ignacio-Souza, L. M., Simino, L. A. P., Milanski, M., Velloso, L. A., Torsoni, M. A., and Torsoni, A. S.: Maternal high-fat diet consumption modulates hepatic lipid metabolism and microRNA-122 (miR-122) and microRNA-370 (miR-370) expression in offspring, Br. J. Nutr., 111, 2112–2122, https://doi.org/10.1017/S0007114514000579, 2014.
Bond, L. M., Miyazaki, M., O'Neill, L. M., Ding, F., and Ntambi, J. M.: Fatty Acid Desaturation and Elongation in Mammals, in: Biochemistry of Lipids, Lipoproteins and Membranes, 6th Edn., Elsevier, 185–208, https://doi.org/10.1016/B978-0-444-63438-2.00006-7, 2016.
Dávalos, A., Goedeke, L., Smibert, P., Ramírez, C. M., Warrier, N. P., Andreo, U., Cirera-Salinas, D., Rayner, K., Suresh, U., Pastor-Pareja, J. C., Esplugues, E., Fisher, E. A., Penalva, L. O. F., Moore, K. J., Suárez, Y., Lai, E. C., and Fernández-Hernando, C.: miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling, P. Natl. Acad. Sci. USA, 108, 9232–9237, https://doi.org/10.1073/pnas.1102281108, 2011.
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., Watts, L., Booten, S. L., Graham, M., McKay, R., Subramaniam, A., Propp, S., Lollo, B. A., Freier, S., Bennett, C. F., Bhanot, S., and Monia, B. P.: miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting, Cell Metab., 3, 87–98, https://doi.org/10.1016/j.cmet.2006.01.005, 2006.
Gerin, I., Bommer, G. T., McCoin, C. S., Sousa, K. M., Krishnan, V., and MacDougald, O. A.: Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis, Am. J. Physiol.-Endocrinol. Metab., 299, 198–206, https://doi.org/10.1152/ajpendo.00179.2010, 2010.
Gerosa, S. and Skoet, J.: Milk availability: Trends in production and demand and medium-term outlook, ESA Work. Pap., 12, 1–40, 2012.
Green, C. D., Ozguden-Akkoc, C. G., Wang, Y., Jump, D. B., and Olson, L. K.: Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species, J. Lipid Res., 51, 1871–1877, https://doi.org/10.1194/jlr.M004747, 2010.
He, M., Kratz, L. E., Michel, J. J., Vallejo, A. N., Ferris, L., Kelley, R. I., Hoover, J. J., Jukic, D., Gibson, K. M., Wolfe, L. A., Ramachandran, D., Zwick, M. E., and Vockley, J.: Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay, J. Clin. Invest., 121, 976–984, https://doi.org/10.1172/JCI42650, 2011.
He, M., Smith, L. D., Chang, R., Li, X., Vockley, J., Feingold, K. R., and Elias, P.: The role of sterol-C4-methyl oxidase in epidermal biology ?, BBA-Molecular Cell Biol. Lipids, 1841, 331–335, https://doi.org/10.1016/j.bbalip.2013.10.009, 2014.
Horie, T., Baba, O., Kuwabara, Y., Yokode, M., Kita, T., and Kimura, T.: MicroRNAs and Lipoprotein Metabolism, J. Atheroscler. Rhrombosis, 21, 17–22, https://doi.org/10.5551/jat.20859, 2014.
Iliopoulos, D., Drosatos, K., Hiyama, Y., Goldberg, I. J., and Zannis, V. I.: MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1α and affects lipid metabolism, J. Lipid Res., 51, 1513–1523, https://doi.org/10.1194/jlr.M004812, 2010.
Iqbal, A., Ziyi, P., Yu, H., Jialing, L., Haochen, W., Jing, F., and Ping, J.: C4BPA: A Novel Co-Regulator of Immunity and Fat Metabolism in the Bovine Mammary Epithelial Cells, Front. Genetics, 12, 1–16, https://doi.org/10.3389/fgene.2021.830566, 2022.
Junjvlieke, Z., Khan, R., Mei, C., Cheng, G., Wang, S., Raza, S. H. A., Hong, J., Wang, X., Yang, W., and Zan, L.: Effect of ELOVL6 on the lipid metabolism of bovine adipocytes, Genomics, 112, 2282–2290, https://doi.org/10.1016/j.ygeno.2019.12.024, 2020.
Kwon, H. J., Kim, S. N., Kim, Y. A., and Lee, Y. H.: The contribution of arachidonate 15-lipoxygenase in tissue macrophages to adipose tissue remodeling, Cell Death Dis., 7, e2285-11, https://doi.org/10.1038/cddis.2016.190, 2016.
Lee, K., Kerner, J., and Hoppel, C. L.: Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex, J. Biol. Chem., 286, 25655–25662, https://doi.org/10.1074/jbc.M111.228692, 2011.
Leonard, A. E., Pereira, S. L., Sprecher, H., and Huang, Y. S.: Elongation of long-chain fatty acids, Prog. Lipid Res., 43, 36–54, https://doi.org/10.1016/S0163-7827(03)00040-7, 2004.
Leroux, C., Bernard, L., Faulconnier, Y., Rouel, J., De La Foye, A., Domagalski, J., and Chilliard, Y.: Bovine mammary nutrigenomics and changes in the milk composition due to rapeseed or sunflower oil supplementation of high-forage or high-concentrate diets, J. Nutrigenet. Nutrigenomics, 9, 65–82, https://doi.org/10.1159/000445996, 2016.
Li, X., Jiang, P., Yu, H., Yang, Y., Xia, L., Yang, R., Fang, X., and Zhao, Z.: MiR-21-3p Targets Elovl5 and Regulates Triglyceride Production in Mammary Epithelial Cells of Cow, DNA Cell Biol., 38, 352–357, https://doi.org/10.1089/dna.2018.4409, 2019.
Lin, X., Luo, J., Zhang, L., and Zhu, J.: MicroRNAs synergistically regulate milk fat synthesis in mammary gland epithelial cells of dairy goats, Gene Expr., 16, 1–13, https://doi.org/10.3727/105221613X13776146743262, 2013.
Lu, C., Yang, R., Liu, B., Li, Z., Shen, B., Yan, S., Zhang, Y., Zhang, L., and Zhao, Z.: Establishment of two types of mammary epithelial cell lilies from Chinese Holstein dairy cow, J. Anim. Vet. Adv., 11, 1166–1172, https://doi.org/10.3923/javaa.2012.1166.1172, 2012.
Lu, F., Luo, C., Li, N., Liu, Q., Wei, Y., Deng, H., Wang, X., Li, X., Jiang, J., Deng, Y., and Shi, D.: Efficient Generation of Transgenic Buffalos (Bubalus bubalis) by Nuclear Transfer of Fetal Fibroblasts Expressing Enhanced Green Fluorescent Protein, Sci. Rep., 8, 1–10, https://doi.org/10.1038/s41598-018-25120-5, 2018.
Matsuzaka, T., Shimano, H., Yahagi, N., Kato, T., Atsumi, A., Yamamoto, T., Inoue, N., Ishikawa, M., Okada, S., Ishigaki, N., Iwasaki, H., Iwasaki, Y., Karasawa, T., Kumadaki, S., Matsui, T., Sekiya, M., Ohashi, K., Hasty, A. H., Nakagawa, Y., Takahashi, A., Suzuki, H., Yatoh, S., Sone, H., Toyoshima, H., Osuga, J. I., and Yamada, N.: Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance, Nat. Med., 13, 1193–1202, https://doi.org/10.1038/nm1662, 2007.
Moon, Y. A., Shah, N. A., Mohapatra, S., Warrington, J. A., and Horton, J. D.: Identification of a Mammalian Long Chain Fatty Acyl Elongase Regulated by Sterol Regulatory Element-binding Proteins, J. Biol. Chem., 276, 45358–45366, https://doi.org/10.1074/jbc.M108413200, 2001.
Moon, Y. A., Ochoa, C. R., Mitsche, M. A., Hammer, R. E., and Horton, J. D.: Deletion of ELOVL6 blocks the synthesis of oleic acid but does not prevent the development of fatty liver or insulin resistance, J. Lipid Res., 55, 2597–2605, https://doi.org/10.1194/jlr.M054353, 2015.
Muir, K., Hazim, A., He, Y., Peyressatre, M., Kim, D. Y., Song, X., and Beretta, L.: Proteomic and lipidomic signatures of lipid metabolism in NASH-associated Hepatocellular carcinoma, Cancer Res., 73, 4722–4731, https://doi.org/10.1158/0008-5472.CAN-12-3797, 2013.
Niu, Y., Zhao, X., Zhou, J., Li, Y., Huang, Y., Cai, B., Liu, Y., Ding, Q., Zhou, S., Zhao, J., Zhou, G., Ma, B., Huang, X., Wang, X., and Chen, Y.: Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9, Reprod. Fertil. Dev., 30, 307–312, https://doi.org/10.1071/RD17068, 2018.
Rayner, K. J., Esau, C. C., Hussain, F. N., Mcdaniel, A. L., Marshall, S. M., Gils, J. M. Van, Ray, T. D., Sheedy, F. J., Goedeke, L., Liu, X., Khatsenko, O. G., Kaimal, V., Lees, C. J., Fernandez-hernando, C., Fisher, E. A., Temel, R. E., and Moore, K. J.: raises plasma HDL and lowers VLDL triglycerides, Nature, 478, 404–407, https://doi.org/10.1038/nature10486, 2011.
Rayner, K. J., Fernández-Hernando, C., and Moore, K. J.: MicroRNAs regulating lipid metabolism in atherogenesis, Thromb. Haemost., 107, 642–647, https://doi.org/10.1160/th11-10-0694, 2012.
Rottiers, V., Obad, S., Petri, A., McGarrah, R., Lindholm, M. W., Black, J. C., Sinha, S., Goody, R. J., Lawrence, M. S., DeLemos, A. S., Hansen, H. F., Whittaker, S., Henry, S., Brookes, R., Najafi-Shoushtari, S. H., Chung, R. T., Whetstine, J. R., Gerszten, R. E., Kauppinen, S., and Näär, A. M.: Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR, Sci. Transl. Med., 5, 1–11, https://doi.org/10.1126/scitranslmed.3006840, 2013.
Shen, B., Zhang, L., Lian, C., Lu, C., Zhang, Y., Pan, Q., Yang, R., and Zhao, Z.: Deep sequencing and screening of differentially expressed microRNAs related to milk fat metabolism in bovine primary mammary epithelial cells, Int. J. Mol. Sci., 17, 2–16, https://doi.org/10.3390/ijms17020200, 2016.
Shen, B., Yang, Z., Han, S., Zou, Z., Liu, J., Nie, L., Dong, W., Li, E., Liu, S., Zhao, Z., and Wu, R.: Bta-miR-124a Affects Lipid Metabolism by Regulating PECR Gene, Biomed Res. Int., 2019, 1–10, 2596914, https://doi.org/10.1155/2019/2596914, 2019.
Shi, H., Zhu, J., Luo, J., Cao, W., Shi, H., Yao, D., Li, J., Sun, Y., Xu, H., Yu, K., and Loor, J. J.: Genes regulating lipid and protein metabolism are highly expressed in mammary gland of lactating dairy goats, Funct. Integr. Genomics, 15, 309–321, https://doi.org/10.1007/s10142-014-0420-1, 2015.
Shi, H. B., Wu, M., Zhu, J. J., Zhang, C. H., Yao, D. W., Luo, J., and Loor, J. J.: Fatty acid elongase 6 plays a role in the synthesis of long-chain fatty acids in goat mammary epithelial cells, J. Dairy Sci., 100, 4987–4995, https://doi.org/10.3168/jds.2016-12159, 2017.
Winckel, M. V., Velde, S. V., Bruyne, R. D., and Biervliet, S. V.: Clinical practice: vegetarian infant and child nutrition, Eur. J. Pediatr., 170, 1489–1494, https://doi.org/10.1007/s00431-011-1547-x, 2011.
Xu, J., Shao, T., Ding, N., Li, Y., and Li, X.: miRNA – miRNA crosstalk: from genomics to phenomics, Brief. Bioinform., 18, 1–10, https://doi.org/10.1093/bib/bbw073, 2016.
Yang, Y., Pan, Q., Sun, B., Yang, R., Fang, X., Liu, X., Yu, X., and Zhao, Z.: miR-29b Targets LPL and TDG Genes and Regulates Apoptosis and Triglyceride Production in MECs, DNA Cell Biol., 35, 758–765, https://doi.org/10.1089/dna.2016.3443, 2016.
Yang, Y., Fang, X., Yang, R., Yu, H., Jiang, P., Sun, B., and Zhao, Z.: MIR-152 Regulates Apoptosis and Triglyceride Production in MECs via Targeting ACAA2 and HSD17B12 Genes, Sci. Rep., 8, 1–10, https://doi.org/10.1038/s41598-017-18804-x, 2018.