the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The expression and mutation of BMPR1B and its association with litter size in small-tail Han sheep (Ovis aries)
Yu-Liang Wen
Xiao-Fei Guo
Lin Ma
Xiao-Sheng Zhang
Jin-Long Zhang
Ming-Xing Chu
Previous studies have shown that BMPR1B promotes follicular development and ovarian granulosa cell proliferation, thereby affecting ovulation in mammals. In this study, the expression and polymorphism of the BMPR1B gene associated with litter size in small-tail Han (STH) sheep were determined. The expression of BMPR1B was detected in 14 tissues of STH sheep during the follicular phase as well as in the hypothalamic–pituitary–gonadal (HPG) axis of monotocous and polytocous STH sheep during the follicular and luteal phases using quantitative polymerase chain reaction (qPCR). Sequenom MassARRAY® single nucleotide polymorphism (SNP) technology was also used to detect the polymorphism of SNPs in seven sheep breeds. Here, BMPR1B was highly expressed in hypothalamus, ovary, uterus, and oviduct tissue during the follicular phase, and BMPR1B was expressed significantly more in the hypothalamus of polytocous ewes than in monotocous ewes during both the follicular and luteal phases (P<0.05). For genotyping, we found that genotype and allele frequencies of three loci of the BMPR1B gene were extremely significantly different (P<0.01) between the monotocous and polytocous groups. Association analysis results showed that the g.29380965A>G locus had significant negative effects on the litter size of STH sheep, and the combination of g.29380965A>G and FecB (Fec – fecundity and B – Booroola; A746G) at the BMPR1B gene showed that the litter size of AG–GG, AA–GG, and GG–GG genotypes was significantly higher compared with other genotypes (P<0.05). This is the first study to find a new molecular marker affecting litter size and to systematically analyze the expression of BMPR1B in different fecundity and physiological periods of STH sheep.
- Article
(747 KB) - Full-text XML
- BibTeX
- EndNote
Lambing is an important economic trait of sheep and is closely related to the economic benefits of sheep breeding. However, as it is a complex threshold trait, it is very inefficient if it relies on traditional breeding methods (Miao and Luo, 2013). The genes affecting the prolificacy of sheep have received much attention from researchers since the 1980s (Tang et al., 2018). Bone morphogenetic protein receptor 1B (BMPR1B), as the receptor of the bone morphogenetic protein (BMP) family, has been identified as a major gene affecting the litter size in sheep (Chu et al., 2011; Wozney et al., 1988; Juengel et al., 2013). Studies have shown that BMPR1B is widely expressed in mammalian ovaries, including humans, mice, sheep, and cattle, and plays an important role in early embryonic development, synthesis of the extracellular matrix, and regulation of ovarian development in sheep (Souza et al., 2001; Elizabeth et al., 2010; Juengel et al., 2013; Khalaf et al., 2013; Selvaraju et al., 2013). Mice deficient in the BMPR1B gene have been shown to experience infertility, as BMPR1B deficiency affects the proliferation of cumulus granulosa cells and reduces the aromatase content, causing irregular estrous cycles (Mishina et al., 1995; Sun and Li, 2013).
BMPR1B is a bone morphogenetic protein type 1 receptor containing 11 exons that encoding 502 amino acids with a coding region of 1509 bp. Its extracellular region contains approximately 150 amino acids, and the intracellular region of the first 30 amino acids of the kinase domain contains a unique GS (glycine- and serine-rich sequence) domain and a 55 ku glycoprotein (Emmerson et al., 2018; Li et al., 2018). FecB (Fec – fecundity and B – Booroola; A746G) missense mutation in the BMPR1B gene causes amino acid changes (Q249R), thereby increasing ovulation and lambing in sheep (Souza et al., 2001). This mutation has an additive effect on ovulation in sheep – that is, the increase in the ovulation increases by one copy (Piper et al., 1985; Guo et al., 2018; Chong et al., 2019). A previous study has shown that BMPR1B mutation increases the signal intensity of the signal transduction process to downstream receptors, leading to premature follicles and increased ovulation (Guo et al., 2018). The FecB mutation is widely distributed. In addition to Booroola Merino sheep, the mutation, which can improve twin production (El Seedy et al., 2017; Darwish, 2018), has been detected in Kendrapada sheep (Mahdavi et al., 2014) in India, Javanese sheep (Kumar et al., 2008) in Indonesia, Kalehkoohi sheep (Mahdavi et al., 2014) in Iran, and Hu sheep (Feng et al., 2006) in China. With deep research into high-fecundity genes in sheep, the FecB gene has been increasingly applied to the cultivation of new sheep varieties. For example, Chen et al. (2015) used the FecB effect to cross small-tail Han (STH) sheep with Dorper sheep, and the average litter size in the hybrid offspring was significantly higher than in Dorper sheep (P<0.05). CRISPR/Cas9 technology has also been applied to sheep embryos, establishing a technical basis for editing the sheep BMPR1B gene (Zhang et al., 2017; Zhou et al., 2018; Rui et al., 2019). Given the significance of the BMPR1B gene, the discovery of other SNPs is particularly important. The whole genomes were re-sequenced in 10 sheep breeds in the early stage of our study. Three SNPs were screened based on the Oar_3.1 reference genome (Pan et al., 2018). Therefore, we hypothesized that these SNPs may be related to the litter size in STH sheep. Thus, the expression pattern of the BMPR1B gene in STH sheep during different fecundity (polytocous and monotocous) and different physiological (follicular and luteal) periods were detected using quantitative polymerase chain reaction (qPCR). The Sequenom MassARRAY® single nucleotide polymorphism (SNP) technique was used to genotype the three SNPs in a large group, and association analysis was then conducted with respect to the litter size of STH sheep. The study focused on the BMPR1B gene expression, new molecular markers affecting litter size, and how these mutations provided new insights into the control of ovarian function in STH sheep.
2.1 Animal processing
A total of 12 STH ewes (FecB genotype) were randomly selected from Yuncheng (Yuncheng County, Shandong, China): six monotocous and six polytocous ewes according to lambing records. All ewes were healthy and approximately 3 years old. The abovementioned 12 test sheep were injected with vaginal progesterone embolization (controlled internal drug releasing, CIDR) for simultaneous estrus. After 12 d, follicular development and ovulation were observed via laparoscopy to determine the estrus period and sampling time; specifically, the follicular phase was 45 h after withdrawal, and the luteal phase was 10 d after withdrawal. A total of 14 tissue samples (heart, liver, spleen, lung, kidney, thyroid, adrenal gland, brain, cerebellum, hypothalamus, pituitary, ovary, uterus, and oviduct) were collected from each six follicular-phase (three monotocous and three polytocous) ewes and six luteal-phase (three monotocous and three polytocous) ewes after the 12 ewes were slaughtered. All selected samples were stored at −80 ∘C for RNA extraction.
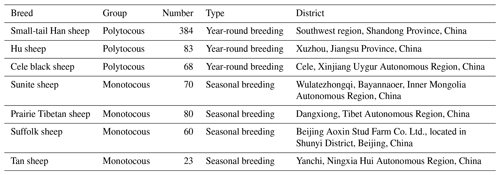
A total of 768 blood samples collected from seven sheep breeds were used for DNA extraction, including three monotocous breeds (384 STH sheep, 83 Hu sheep, and 68 Cele black sheep) and four polytocous breeds (80 Prairie Tibetan sheep, 60 Suffolk sheep, 70 Sunite sheep, and 23 Tan sheep) (Table 1).
2.2 Total DNA and RNA preparation
Blood DNA and tissue RNA were extracted using a DNA extraction kit (TIANGEN, Beijing, China) and an RNA extraction kit (TIANGEN, Beijing, China), respectively, with TRIzol (Invitrogen Inc., Carlsbad, CA, USA). The quantity and quality of total DNA and RNA were monitored to ensure that they met the requirements for subsequent experiments; the reader is referred to Chen et al. (2020) for details on the specific methods utilized.
2.3 Primer design
The primers for the sheep BMPR1B and RPL19 genes were designed based on their sequences in GenBank (Table 2). RPL19 (accession no. XM_012186026.1) was used as an internal control to normalize the threshold cycle (Ct) values. Primers were synthesized by Beijing Tianyi Biotechnology (Beijing, China).
2.4 qPCR
The first strand of cDNA was prepared following the instructions of the PrimeScript™ RT Reagent Kit (TaKaRa Bio Inc., Dalian, China). Real-time quantitative polymerase chain reaction (qPCR) amplification was performed in a 20 µL reaction mixture, containing 10 µL SYBR Premix (TaKaRa, Dalian, China), 0.4 µL of each forward and reverse primer (20 ng/µL), 7.2 µL ddH2O, and 2 µL cDNA (200 ng/µL). The reaction conditions were as follows: initial denaturation at 95 ∘C for 5 min, denaturation at 95 ∘C for 10 s, annealing at 60 ∘C for 30 s, and extension at 72 ∘C for 30 s (40 cycles). The relative expression levels of BMPR1B and RPL19 mRNA were analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.5 Genotyping
In the early stage of our study, three SNPs (g.29380950G>A, g.29380965A>G, and g.29401381C>T) of the BMPR1B gene were obtained. The above SNPs were then typed using the Sequenom MassARRAY® SNP technique. The 768 blood DNA (40–80 ng/µL, 20 µL) samples from the different breeds were used for genotyping (Table 1); the reader is referred to Zhou et al. (2018) for details on the typing step utilized.
2.6 Statistical analysis
Statistical analyses were performed using an analysis of variance (ANOVA), followed by a Fisher's least significant difference test as a multiple comparison test in SPSS 19.0 software (IBM, Armonk, New York, USA). The adjusted linear model, yijkl=μ + Genotypei + Sk+eijkl, in which yijkl is the trait observation, μ is the overall average, Genotypei is the genotype effect, Sk is the season effect, and eijkl is the random error, was applied to determine the association between genotypes and the litter size in STH sheep. It is assumed that random errors (eijkl) are independent of each other and obey the N(0,σ2) distribution. The allele and genotype frequency, polymorphism information content (PIC), heterozygosity (He), number of effective alleles (Ne), and χ2 (chi-square) value were calculated using the data from genotyping; ewe populations with P>0.05 (based on the χ2 test) were considered to be in Hardy–Weinberg equilibrium. All experimental data are presented as mean ± SE (standard error of the mean): P≤0.05 indicates that the difference was significant; P≤0.01 indicates that the difference was extremely significant.
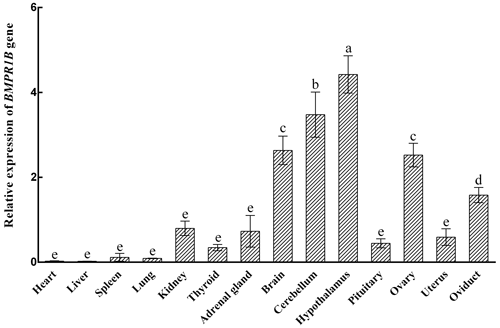
3.1 Expression levels of BMPR1B in different tissues during the follicular phase
The expression characteristics of the BMPR1B gene are shown in Fig. 1. The BMPR1B gene was highly expressed in the brain and in reproduction-related tissues. Taking the expression of the BMPR1B gene in the pituitary as a reference, the expression levels in the brain, cerebellum, hypothalamus, ovary, and oviduct were 5.92 times (P<0.001), 7.82 times (P<0.001), 9.95 times (P<0.001), 5.67 times (P<0.001), and 3.56 times (P=0.004) higher, respectively. The expression of BMPR1B in the uterus was 1.33 times higher than that of pituitary tissue, but this difference was not significant (P>0.05).
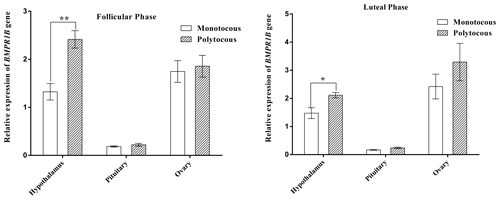
3.2 Expression of BMPR1B in the HPG axis tissues during different physiological periods
As shown in Fig. 2, BMPR1B expression in the hypothalamus of the polytocous group in the follicular and luteal phases was significantly higher than expression in the monotocous group (P<0.05). The expression of the polytocous group in the pituitary and ovarian tissues in the follicular and luteal phases was higher than that of the monotocous group but did not reach a significant level (P>0.05).
3.3 Polymorphism of the BMPR1B gene
The genotyping results of the g.29380950G>A, g.29380965A>G, and g.29401381C>T loci of BMPR1B are shown in Fig. 3. For g.29380950G>A (Fig. 3a), for example, there are three genotypes: wild homozygous AA, heterozygous GA, and mutant homozygous GG. For the g.29401381C>T locus, in contrast, there are only two genotypes: homozygous CC and heterozygous CT (Fig. 3c).
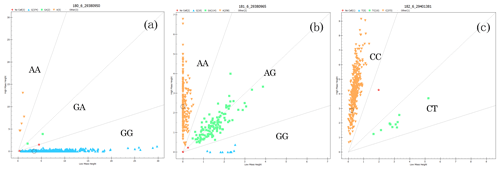
Figure 3Genotyping in three loci of BMPR1B. The upper-left area indicates the AA genotype (a) with 5 individuals, the AA genotype (b) with 258 individuals, and the CC genotype (c) with 373 individuals; the middle area indicates the GA genotype (a) with 2 individuals, the AG genotype (b) with 114 individuals, and the CT genotype (c) with 10 individuals; the lower-right area indicates the GG genotype (a) with 374 individuals and the GG genotype (b) with 10 individuals.
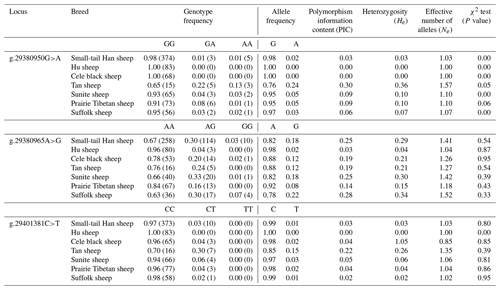
3.4 Population genetic analysis of SNPs in the BMPR1B gene
Three SNPs (g.29380950G>A, g.29380965A>G, and g.29401381C>T) in BMPR1B were detected in monotocous and polytocous sheep breeds (Table 3) and for seven sheep breeds (Table 4). The allele frequencies of the g.29380950G>A, g.29380965A>G, and g.29401381C>T loci between monotocous and polytocous breeds were significantly different (P<0.01). The difference in the genotype frequency of the g.29380950G>A locus between monotocous and polytocous sheep breeds reached an extremely significant level (P<0.001), and the difference in the genotype frequency of the g.29401381C>T locus reached a significant level (P<0.05). The difference in the g.29380965A>G locus genotype frequency between monotocous and polytocous sheep breeds, in contrast, did not reach a significant level (P=0.97).
Table 3Genotype and allele frequencies of three SNPs of the BMPR1B gene in monotocous and polytocous sheep.
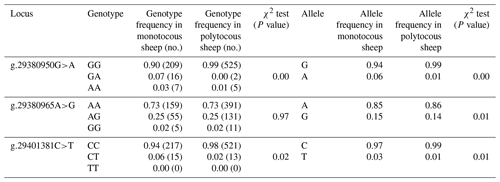
P<0.05 denotes significant differences, and P<0.01 denotes extremely significant differences.
The allele frequency, genotype frequency, PIC, He, Ne, and χ2 test results from population genetic analysis for three SNPs in the seven sheep breeds are listed in Table 4. The g.29380950G>A locus showed moderate polymorphism (0.25 < PIC < 0.5) in the Tan sheep breed and low polymorphism in the other sheep breeds (PIC < 0.25); the g.29380965A>G locus showed moderate polymorphism (0.25 < PIC < 0.5) in STH sheep, Sunite sheep, and Suffolk sheep, whereas it displayed low polymorphism in other breeds (PIC < 0.25); the g.29401381C>T locus showed low polymorphism in all seven sheep breeds (PIC < 0.25). The χ2 test results showed that the g.29380950G>A locus was not in Hardy–Weinberg equilibrium (P<0.05) except for in the Tan and Prairie Tibetan sheep breeds; the g.29380965A>G locus was in Hardy–Weinberg equilibrium in the seven sheep breeds (P>0.05); the g.29401381C>T locus was in Hardy–Weinberg equilibrium in all sheep breeds (P>0.05) except for Hu sheep.
3.5 Association between three loci of BMPR1B and litter size in small-tail Han sheep
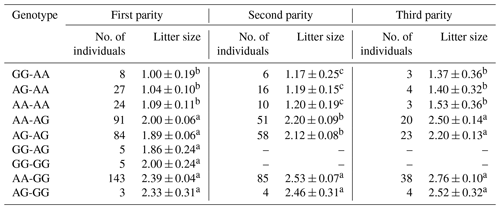
The results revealed that the g.29380965A>G locus was significantly correlated with litter size in STH sheep (Table 5) and that the litter size of ewes with AA and AG genotypes was higher than that of ewes with the GG genotype (P<0.05). As for the combination of the g.29380965A>G locus and the FecB genotype (Table 6), the composite genotype was significantly correlated with litter size in STH sheep (P<0.05): the litter sizes of ewes with AG–GG, AA–GG, and GG–GG genotypes were significantly higher than other combination genotypes (P<0.05).
Table 5Analysis of different genotypes of BMPR1B and litter size in small-tail Han sheep.
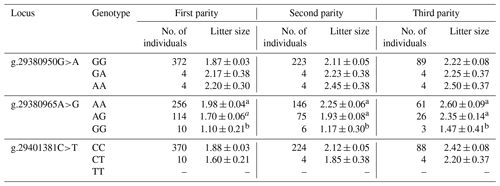
Note that different lowercase letters in the same column represent a significant difference (P<0.05).
4.1 Effect of BMPR1B gene expression on mammalian reproduction
BMPR1B (FecB) is one of the major fecundity genes in female reproduction and plays a major role in the development of follicles and the proliferation of ovarian granulosa cells in sheep (Shimasaki et al., 1999; Chu et al., 2007; Yao et al., 2019; Tao et al., 2019; Abdurahman et al., 2019). Previous reports have found that BMPR1B belongs to the type I receptors of BMPs (Aquino et al., 2017), which figure prominently in the proliferation of primordial germ cells (PGCs) (Nermin et al., 2018; Yi et al., 2001) and in the production and secretion of reproductive hormone (Richards and Pangas, 2010; Isaacs et al., 1995) by binding specifically to different types of ligands (Ikeda et al., 2016). Previous studies have found that BMPR1B is widely expressed in the brain and reproduction-related tissues of mammals (Goyal et al., 2017; Foroughinia et al., 2017) and is moderately expressed in heart, liver, spleen, lung, and muscle tissue (Tang et al., 2018; Ciller et al., 2016). In this study, BMPR1B was found to be expressed in all selected STH tissue types and was highly expressed in the brain, cerebellum, hypothalamus, ovary, and oviduct. This shows that it may play a major role in the normal physiological functions of various organs.
The expression of BMPR1B in the hypothalamic–pituitary–gonadal (HPG) axis was higher in the polytocous group than in the monotocous group in the follicle and luteal phases, reaching a significant level in the hypothalamus in the follicle phase and an extremely significant level in the luteal phase. This observation was identical to a previous study comparing high-fecundity and low-fecundity ewes (Xu et al., 2010; Goyal et al., 2017). Tang et al. (2018) explored the expression of BMPR1B in the HPG axis tissues using three genotypes (BB, B+, and genotypes of FecB) and found that BMPR1B expression was significantly higher in the hypothalamus and pituitary in the BB group compared with the B+ and groups. The BMPR1B gene has also been extensively studied in other species and was found to be more highly expressed in the ovary of a highly prolific goat breed (Jintang black goat) than in that of a breed with a low reproductive output (Tibetan goat) (Pan et al., 2015). In terms of embryonic development, BMPR1B has been found to play an important role in mouse embryonic development, and BMPR1B defects in mice have been shown to cause infertility (Mishina et al., 1995). In addition, BMPR1B defects affect the proliferation of cumulus granulosa cells and reduce the aromatase content, causing mice to exhibit an irregular estrous cycle (Yi et al., 2001; Sun and Li, 2013).
BMPR1B also plays an important role in follicular development as well as the synthesis and secretion of reproductive hormones. In buffalo ovarian follicular cells, according to the diameter of the follicle and the content of E2 (Estradiol), the follicle cells are divided into dominant follicles (D > 13 mm, E2>180 ng/mL) and other cells, and the expression of BMPR1B in the granulosa cells and ovarian membrane cells of dominant follicles is 1.5–2.0 times higher than that of other follicles (Rajesh et al., 2018; Elizabeth et al., 2010). Costa et al. (2012) found that the expression of the BMPR1B gene in a 0.2 mm follicle was higher than that in a 1.0 mm follicle in the goat ovary. For reproductive hormones, the expression of BMPR1B was directly proportional to the increase in E2 content (Paradis et al., 2009). A large number of studies have shown that the BMPR1B gene plays an active role in mammalian embryo development and follicular development (Tang et al., 2019; Yao et al., 2019; Abdurahman et al., 2019; Zhao et al., 2016). However, further studies are required to deeply investigate the relationship between BMPR1B and mammalian reproduction.
4.2 Relationship between BMPR1B mutation and litter size in sheep
According to their analysis of litter size records from Booroola Merino sheep, Piper and Bindon (1982) reported that the exceptional fecundity of this breed may in part result from the action of a single major gene affecting the ovulation rate. In 1989, the Sheep and Goat Genetic Nomenclature Committee termed this gene “FecB” (fecundity Booroola). This point mutation A746G was in the coding region of the BMPR1B gene (Souza et al., 2001). To date, the BMPR1B mutation (FecB) has been found in various sheep breeds, such as Garole sheep (India; Davis et al., 2002; Polley et al., 2010), Javanese sheep (Indonesia; Davis et al., 2002), and Bayanbulak sheep (China; Zuo et al., 2013). Moreover, the polymorphism of the BMPR1B gene has also been extensively studied in other species. Darwish (2018) found that BMPR1B polymorphism was significantly associated with litter size in Egyptian goats, which could significantly increase the rate of twin births. By constructing a vector with the mutant G allele of the A746G mutation, Zhao et al. (2016) found that BMPR1B expression in pigs was 0.5–2 times higher in multiple tissue types of transgenic-positive F1 piglets than in their transgenic-negative siblings. Moreover, with further study of BMPR1B, more polymorphic sites have been found: Dutta et al. (2014) found a new mutation G773C in the Assam hill goat that may have a completely different mutation effect to A746G (FecB). Furthermore, mutation g.66496G>A in exon 8 of the BMPR1B was found in prolific Lori-Bakhtiari ewes and was reported to be significantly related to litter size (Abdoli et al., 2018).
BMPR1B has been identified as a major gene affecting the litter size in sheep (Juengel et al., 2013; Piper et al., 1985; Davis et al., 1982). Therefore, determining other SNPs is of great significance for improving sheep reproductive performance. In this study, we found that the allele frequencies of the g.29380950G>A, g.29380965A>G, and g.29401381C>T loci was significantly different between monotocous and polytocous breeds; this means that the three loci are significantly related to lambing traits. Additionally, most ewes contained three genotypes: two g.29401381C>T-locus-only genotypes were detected in all breeds, which may have resulted from low polymorphism, and three genotypes were detected if we expanded the number of ewes examined. In addition, the three loci were not in Hardy–Weinberg equilibrium (P<0.05) in some breeds, such as g.29380950G>A in all sheep breeds except for Tan and Prairie Tibetan sheep and g.29401381C>T in the Hu sheep breed; this may have resulted from both natural and artificial selection.
Each copy of the FecB mutation was found to increase ovulation by 1.65 in the highly prolific Booroola Merino breed (Piper et al., 1985). This may increase the signal intensity of the signal transduction process to downstream receptors, leading to premature follicles, increased ovulation, and a subsequent increase in the litter size (Guo et al., 2018). In this study, the g.29380965A>G locus shows similar mutations that do not cause amino acid change but have a significant negative correlation with litter size in STH sheep. The statistics regarding the litter size of the three genotypes showed that the litter size associated with the GG genotype was significantly lower than that associated with the AA and AG genotypes. Association analysis between the combined genotypes of the g.29380965A>G locus and FecB and litter size in STH sheep showed that the liter sizes associated with the AG–GG, AA–GG, and GG–GG genotypes were significantly higher than that associated with other genotypes; this may be due to the locus attenuating the downstream signal intensity of the BMP/SMAD signaling pathway, thereby weakening the effect of the FecB gene in the ovary of STH sheep, reducing ovulation, and decreasing the litter size. However, further studies are required to deeply investigate the molecular mechanism at the cellular level. Nevertheless, this study indicated that the g.29380965A>G locus may be the key locus affecting the litter size of STH sheep; thus, it could be used for selective breeding for an increased litter size in sheep.
In this study, we found that the BMPR1B gene was mainly expressed in the brain, cerebellum, hypothalamus, ovary, uterus, and oviduct of STH sheep and that the expression in the hypothalamus of the polytocous group was significantly higher than that in the monotocous group. We analyzed the population genetics of three SNPs in BMPR1B using association analysis and found that one key locus (g.29380965A>G) can significantly reduce the liter size in STH sheep. Further analysis indicated that this locus influenced litter size in ewes by effecting FecB.
Experimentation on the sheep was conducted according to the Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS; Beijing, China). Ethical approval was provided by the animal ethics committee of IAS-CAAS (no. IAS2019-49, 18 September 2019).
The data sets used in this paper are available from the corresponding author upon request.
These studies were designed by YLW and MXC, who performed the experimental analyses and prepared the figures and tables. YLW analyzed the data and drafted the paper. MXC contributed to revisions of the paper. XFG and LM conducted the background investigation. XSZ, JLZ, and SGZ developed the methodology. All authors read and approved the final paper for publication.
The authors declare that they have no conflict of interest.
This research was funded by the National Natural Science Foundation of China (grant nos. 31861143012 and 31772580), the Natural Science Foundation of Tianjin (grant no. 20JCQNJC00630), the earmarked fund for China Agriculture Research System (grant no. CARS-38), the Agricultural Science and Technology Innovation Program of China (grant nos. CAAS-ZDRW202106 and ASTIP-IAS13), the China High-level Talents Special Support Plan Scientific and Technological Innovation Leading Talents Program (grant no. W02020274), and the Tianjin Agricultural Science and Technology Achievements Transformation and Popularization Program (grant no. 201704020).
This paper was edited by Steffen Maak and reviewed by Xingbo Zhao and one anonymous referee.
Abdoli, R., Mirhoseini, S. Z., Hossein Zadeh, N. G., and Zamani, P.: Screening for Causative Mutations of Major Prolificacy Genes in Iranian Fat-Tailed Sheep, Int. J. Fertil. Steril., 12, 51–55, https://doi.org/10.22074/ijfs.2018.5247, 2018.
Abdurahman, A., Du, X., Yao, Y. L., Sulaiman, Y. M., Aniwashi, J., and Li, Q. F.: Smad4 Feedback Enhances BMPR1B Transcription in Ovine Granulosa Cells. Int. J. Mol. Sci., 20, 2732, https://doi.org/10.3390/ijms20112732, 2019.
Aquino Martínez, R., Artigas, N., Gámez, B., Rosa, J. L., and Ventura, F.: Extracellular calcium promotes bone formation from bone marrow mesenchymal stem cells by amplifying the effects of BMP-2 on SMAD signalling, PLoS ONE, 12, e178158, https://doi.org/10.1371/journal.pone.0178158, 2017.
Chen, W. H., Tian, Z. L., Ma, L., Gan, S. Q., Sun, W., and Chu, M. X.: Expression analysis of BMPR1B, BMP15, GDF9, Smad1, Smad5, and Smad9 in rams with different fecundity, Pak. J. Zool., 52, 1665–1674, https://doi.org/10.17582/journal.pjz/20190226030219, 2020.
Chen, X. Y., Sun, H. X., Tian, S. J., Xiang, H., Zhou, L. S., Dun, W. T., and Zhao, X. B.: Increasing litter size in a sheep breed by marker-assisted selection of BMPR1B A746G mutation, J. Genet., 94, 139–142, https://doi.org/10.1007/s12041-015-0470-8, 2015.
Chong, Y. Q., Liu, G. Q., and Jiang, X. P.: Effect of BMPRIB gene on litter size of sheep in China: A meta-analysis, Anim. Reprod. Sci., 210, 106175, https://doi.org/10.1016/j.anireprosci.2019.106175, 2019.
Chu, M. X., Liu, Z. H., Jiao, C. L., He, Y. Q., Fang, L., Ye, S. C., Chen, G. H., and Wang, J. Y.: Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries), J. Anim. Sci., 85, 598–603, https://doi.org/10.2527/jas.2006-324, 2007.
Chu, M. X., Jia, L. H., Zhang, Y. J., Jin, M., Chen, H. Q., Fang, L., Di, R., Cao, G. L., Feng, T., Tang, Q. Q., Ma, Y. H., and Li, K.: Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep, Mol. Biol. Rep., 38, 4071–4076, https://doi.org/10.1007/s11033-010-0526-z, 2011.
Ciller, I. M., Palanisamy, S. K., Ciller, U. A., and Mcfarlane, J. R.: Postnatal expression of bone morphogenetic proteins and their receptors in the mouse testis, Physiol. Res., 65, 673–682, https://doi.org/10.33549/physiolres.933193, 2016.
Costa, J. J., Passos, M. J., Leitão, C. C., Vasconcelos, G. L., Saraiva, M. V., Figueiredo, J. R., Van, D. H. R., and Silva, J. R.: Levels of mRNA for bone morphogenetic proteins, their receptors and SMADs in goat ovarian follicles grown in vivo and in vitro, Reprod. Fertil. Dev., 24, 723–732, https://doi.org/10.1071/rd11195, 2012.
Darwish, H.: Study of genetic polymorphism in BMPRIB gene and its association with improving twin production in Egyptian sheep and goats, J. Anim. Vet. Adv., 10, 1–5, https://doi.org/10.22587/rjavs.2018.10.1.1, 2018.
Davis, G. H., Montgomery, G. W., Allison, A. J., Kelly, R. W., and Bray, A. R.: Segregation of a major gene influencing fecundity in progeny of Booroola Sheep, New. Zeal. J. Agr. Res., 25, 525–529, https://doi.org/10.1080/00288233.1982.10425216, 1982.
Davis, G. H., Galloway, S. M., Ross, I. K., Gregan, S. M., Ward, J., Nimbkar, B. V., Ghalsasi, P. M., Nimbkar, C., Gray, G. D., Subandriyo., Inounu, I., Tiesnamurti, B., Martyniuk, E., Eythorsdottir, E., Mulsan, T. P., Lecerf, F., Hanrahan, J. P., Bradford, G. E., and Wilson, T.: DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation, Biol. Reprod., 66, 1869–1874, https://doi.org/10.1095/biolreprod66.6.1869, 2002.
Dutta, R., Laskar, S., Borah, P., Kalita, D., Das, B., Zaman, G., Barman, N. N., and Saikia, D. P.: Polymorphism and nucleotide sequencing of BMPR1B gene in prolific Assam hill goat, Mol. Biol. Rep., 41, 3677–3681, https://doi.org/10.1007/s11033-014-3232-4, 2014.
El Seedy, A. S., Hashem, N. M., El Azrak, K. M., El Din, A. N., and Salem, M. H.: Genetic screening of FecB, FecXG and FecXI mutations and their linkage with litter size in Barki and Rahmani sheep breeds, Reprod. Domest. Anim., 52, 1133–1137, https://doi.org/10.1111/rda.13002, 2017.
Elizabeth, C. L., Gwenn, A. L. C., Béatrice, M. P., Kadler, K. E., and Stéphane F.: First evidence of bone morphogenetic protein 1 expression and activity in sheep ovarian follicles, Biol. Reprod., 83, 138–146, https://doi.org/10.1095/biolreprod.109.082115, 2010.
Emmerson, P. J., Duffin, K. L., and Wu, X.: GDF15 and growth control, Front. Physiol., 9, 1712, https://doi.org/10.3389/fphys.2018.01712, 2018.
Feng, G., Liu, S. R., Shi, G. Q., Ai, J. T., Mao, D. G., and Yang, L. G.: Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development (in Chinese), Yi. Chuan. Xue. Bao., 33, 117–124, 2006.
Foroughinia, G., Fazileh, A., and Eghbalsaied, S.: Expression of genes involved in BMP and estrogen signaling and AMPK production can be important factors affecting total number of antral follicles in ewes, Theriogenology, 91, 36–43, https://doi.org/10.1016/j.theriogenology.2016.12.023, 2017.
Goyal, S., Aggarwal, J., Dubey, P. K., Mishra, B. P., Ghalsasi, P., Nimbkar, C., Joshi, B. K., and Kataria, R. S.: Expression analysis of genes associated with prolificacy in FecB carrier and noncarrier Indian sheep, Anim. Biotechnol., 28, 220–227, https://doi.org/10.1080/10495398.2016.1262869, 2017.
Guo, X. F., Wang, X. Y., Di, R., Liu, Q. Y., Hu, W. P., He, X. Y., Yu, J. R., Zhang, X. S., Zhang, J. L., Broniowska, K., Chen, W., Wu, C. X., and Chu, M. X.: Metabolic effects of FecB gene on follicular fluid and ovarian vein serum in sheep (Ovis aries), Int. J. Mol. Sci., 19, 539, https://doi.org/10.3390/ijms19020539, 2018.
Ikeda, Y., Hasegawa, A., Tsubamoto, H., Wakimoto, Y., Kumamoto, K., and Shibahara, H.: Effects of gremlin-2 on the transition of primordial follicles during early folliculogenesis in the human ovary, Eur. J. Obstet. Gynecol. Reprod. Biol., 203, 72–77, https://doi.org/10.1016/j.ejogrb.2016.05.026, 2016.
Isaacs, K. L., Mcnatty, K. P., Condell, L., Shaw, L., Heath, D. A., Hudson, N. L., Littlejohn R. P., and Mc Leod, B. J.: Plasma FSH, LH and immunoreactive inhibin concentrations in FecBB/FecBB and FecB+/FecB+ Booroola ewes and rams from birth to 12 months of age, J. Reprod. Fertil., 103, 89–97, https://doi.org/10.1530/jrf.0.1030089, 1995.
Juengel, J. L., Davis, G. H., and McNatty, K. P.: Using sheep lines with mutations in single genes to better understand ovarian function, Reproduction, 146, R111–R123, https://doi.org/10.1530/rep-12-0509, 2013.
Kumar, S., Mishra, A., Kolte, A., Dash, S. K., and Karim, S. A.: Screening for Booroola (FecB) and Galway (FecXG) mutations in Indian sheep, Small. Ruminant. Research., 80, 57–61, https://doi.org/10.1016/j.smallrumres.2008.09.007, 2008.
Li, Q. Q., Du, X., Pan, Z. X., Zhang, L. F., and Li, Q. F.: The transcription factor SMAD4 and miR-10b contribute to E2 release and cell apoptosis in ovarian granulosa cells by targeting CYP19A1, Mol. Cell. Endocrinol., 476, 84-95, https://doi.org/10.1016/j.mce.2018.04.012, 2018.
Livak, K. J. and Schmittgen, T. D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method, Methods., 25, 402–408, https://doi.org/10.1006/meth.2001.1262, 2001.
Mahdavi, M., Nanekarani, S., and Hosseini, S. D.: Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep, Anim. Reprod. Sci., 147, 93–98, https://doi.org/10.1016/j.anireprosci.2014.04.003, 2014.
Miao, X. Y. and Luo, Q. M.: Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing, Reproduction, 145, 587–596, https://doi.org/10.1530/rep-12-0507, 2013.
Mishina, Y., Suzuki, A., Ueno, N., and Behringer, R. R.: Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis, Genes. Dev., 9, 3027–3037, https://doi.org/10.1101/gad.9.24.3027, 1995.
Khalaf, M., Morera, J., Bourret, A., Reznik, Y., Denoual, C., Herlicoviez, M., Mittre, H., and Benhaim, A.: BMP system expression in GCs from polycystic ovary syndrome women and the in vitro effects of BMP4, BMP6, and BMP7 on GC steroidogenesis, Eur. J. Endocrinol., 168, 437–444, https://doi.org/10.1530/eje-12-0891, 2013.
Nermin, E. H., Kandil, O. M., Shawky, A. E. M. A., Al Tohamy, A. F. M., El Sayd, Y. A., Hamdy, A. S., Abou Fandoud, E. S. I., Abdel Azeem, S. N., Abdon, E. R. A. H., and Abdoon, A. S. S.: Investigating the effect of GDF9 , BMP15 , BMP6 and BMPR1B polymorphisms on Egyptian sheep fecundity and their transcripts expression in ovarian cells, Small. Ruminant. Res., 165, 34-40, https://doi.org/10.1016/j.smallrumres.2018.06.010, 2018.
Pan, Z. Y., Di, R., Tang, Q. Q., Jin, H. H., Chu, M. X., Huang, D. W., He, J. N., Liu, Q. Y., Hu, W. P., and Wang, X. Y.: Tissue-specific mRNA expression profiles of GDF9, BMP15, and BMPR1B genes in prolific and non-prolific goat breeds, Czech. J. Anim. Sci., 60, 452–458, https://doi.org/10.1002/mrd.21386, 2015.
Pan, Z. Y., Li, S. D., Liu, Q. Y., Wang, Z., Zhou, Z. K., Di, R., Miao, B. P., Hu, W. P., Wang, X. Y., Hu, X. X., Xu, Z., Wei, D. L., He, X. Y., Yuan, L., Guo, X. F., Liang, B. M., Wang, R. C., Li, X. Y., Cao, X. H., Dong, X. L., Xia, Q., Shi, H. C., Hao, G., Yang, J., Luo, S. C., Zhao, Y. Q., Jin, M., Zhang, Y. J., Lv, S. J., Li, F. K., Ding, G. H., Chu, M. X., and Li, X. Y.: Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization, GigaScience, 7, 1–15, https://doi.org/10.1093/gigascience/giy019, 2018.
Paradis, F., Novak, S., Murdoch, G. K., Dyck, M. K., Dixon, W. T., and Foxcroft, G. R.: Temporal regulation of BMP2, BMP6, BMP15, GDF9, BMPR1A, BMPR1B, BMPR2 and TGFBR1 mRNA expression in the oocyte, granulosa and theca cells of developing preovulatory follicles in the pig, Reproduction, 138, 115–129, https://doi.org/10.1530/rep-08-0538, 2009.
Piper, L. R. and Bindon, B. M.: The Booroola Merino and the performance of medium non-Peppin crosses at Armidale, in: The Booroola Merino, edited by: Piper, L. R., Bindon, B. M., and Nethery, R. D., CSIRO, Melbourne, Australia, 9–20, 1982.
Piper, L. R., Bindon, B. M., and Davis, G. H.: The single gene inheritance of the high litter size of the Booroola Merino, Genet. Reprod. In. Sheep., Butterworths, London, 115–125, https://doi.org/10.1016/B978-0-407-00302-6.50016-7, 1985.
Polley, S., De, S., Brahma, B., Mukherjee, A., Vinesh, P. V., Batabyal, S., Arora, J. S., Pan, S., Samanta, A. K., Datta, T. K., and Goswami, S. L.: Polymorphism of BMPR1B, BMP15 and GDF9 fecundity genes in prolific Garole sheep, Trop. Anim. Health. Prod., 42, 985–993, https://doi.org/10.1007/s11250-009-9518-1, 2010.
Rajesh, G., Mishra, S. R., Paul, A., Punetha, M., Vidyalashmi, G. M., Narayanan, K., Bag, S., Bhure, S. K., Chouhan, V. S., and Maurya, V. P.: Transcriptional and translational abundance of bone morphogenetic protein (BMP) 2, 4, 6, 7 and their receptors BMPR1A, 1B and BMPR2 in buffalo ovarian follicle and the role of BMP4 and BMP7 on estrogen production and survival of cultured granulosa cells, Res. Vet. Sci., 118, 371–388, https://doi.org/10.1016/j.rvsc.2018.04.002, 2018.
Richards, J. S. and Pangas, S. A.: The ovary: basic biology and clinical implications, J. Clin. Invest., 120, 963–972, https://doi.org/10.1172/jci41350, 2010.
Selvaraju, S., Folger, J. K., and Gupta, P.: Stage-specific expression and effect of bone morphogenetic protein 2 on bovine granulosa cell estradiol production: regulation by cocaine and amphetamine regulated transcript, Domest. Anim. Endocrinol., 44, 115–120, https://doi.org/10.1016/j.domaniend.2012.10.002, 2013.
Shimasaki, S., Zachow, R. J., Li, D., Kim, H., Lemura, S., Ueno, N., Sampath, K., Chang, R. J., and Erickson, G. F.: A functional bone morphogenetic protein system in the ovary, P. Natl. Acad. Sci. USA, 96, 7282–7287, https://doi.org/10.1073/pnas.96.13.7282, 1999.
Souza, C., Macdougall, C. C., Campbell, B., McNeilly, A., and Baird, D.: The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene, J. Endocrinol., 169, R1–R6, https://doi.org/10.1677/joe.0.169r001, 2001.
Sun, J. and Li, X. D.: Growth and antrum formation of bovine primary follicles in long-term culture in vitro, Reprod. Biol., 13, 221–228, https://doi.org/10.1016/j.repbio.2013.06.003, 2013.
Tang, J. S., Hu, W. P., Di, R., Liu, Q. Y., Wang, X. Y., Zhang, X. S., Zhang, J. L., and Chu, M. X.: Expression analysis of the prolific candidate genes, BMPR1B, BMP15, and GDF9 in Small Tail Han ewes with three fecundity (FecB gene) genotypes, Animals (Basel), 8, 166, https://doi.org/10.3390/ani8100166, 2018.
Tang, J. S., Hu, W. P., Di, R., Wang, X. Y., Zhang, X. S., Zhang, J. L., Liu, Q. Y., and Chu, M. X.: Expression analysis of BMPR1B, BMP15, and GDF9 in prolific and non-prolific sheep breeds during the follicular phase, Czech. J. Anim. Sci., 64, 439–447, https://doi.org/10.17221/101/2018-CJAS, 2019.
Tao, L., Guo, X. F., Wen, Y. L., Di, R., Liu, Q. Y., Hu, W. P., Zhang, X. S., Zhang, J. L., Wang, X. Y., and Chu, M. X.: Effects of FecB mutation on follicular development and expression of AMH and AMHR2 genes in Small Tail Han sheep (Ovis aries), Journal of Agricultural Biotechnology, 27, 684–691, 2019 (in Chinese).
Wozney, J. M., Rosen, V., Celeste, A. J., Mitsock, L. M., Whitters, M. J., Kriz, R. W., Hewick, R. M., and Wang, E. A.: Novel regulators of bone formation: molecular clones and activities, Science, 242, 1528–1534, https://doi.org/10.1126/science.3201241, 1988.
Xu, Y. F., Li, E. L., Han, Y. D., Chen, L., and Xie, Z. A.: Differential expression of mRNAs encoding BMP/SMAD pathway molecules in antral follicles of high- and low-fecundity Hu sheep, Anim. Reprod. Sci., 120, 47–55, https://doi.org/10.1016/j.anireprosci.2010.02.009, 2010.
Yao, Y. L., Reheman, A., Xu, Y. F., and Li, Q. F.: miR-125b contributes to ovarian granulosa cell apoptosis through targeting BMPR1B, a major gene for sheep prolificacy, Reprod. Sci., 26, 295–305, https://doi.org/10.1177/1933719118770544, 2019.
Yi, S. E., Lapolt, P. S., Yoon, B. S., Chen, J. Y., Lu, J. K., and Lyons, K. M.: The type I BMP receptor BmprIB is essential for female reproductive function, P. Natl. Acad. Sci. USA, 98, 7994–7999, https://doi.org/10.1073/pnas.141002798, 2001.
Zhang, X. M., Li, W. R., Wu, Y. S., Peng, X. R., Lou, B., Wang, L. Q., and Liu, M. J.: Disruption of the sheep BMPR-IB gene by CRISPR/Cas9 in in vitro-produced embryos, Theriogenology, 91, 163–172, https://doi.org/10.1016/j.theriogenology.2016.10.025, 2017.
Zhao, X. Y., Yang, Q., Zhao, K. E., Jiang, C., Ren, D. R., Xu, P., Liao, R. R., Jiang, K., Ma, J. W., Xiao, S. J., Ren, J., and Xing, Y. Y.: Production of transgenic pigs with an introduced missense mutation of the bone morphogenetic protein receptor type IB gene related to prolificacy, Asian-Australas. J. Anim. Sci., 29, 925–937, https://doi.org/10.5713/ajas.15.0505, 2016.
Zhou, M., Cao, X. H., He, X. Y., Sun, Q., Di, R., Hu, W. P., Wang, X. Y., Zhang, X. S., Zhang, J. L., Liu, Q. Y., and Chu, M. X.: Tissue expression and polymorphism of sheep (Ovis aries) FSTL1 gene and their association with litter size (in Chinese), Journal Of Agricultural Biotechnology, 26, 801–810, 2018.
Zhou, S. W., Yu, H. H., Zhao, X. E., Cai, B., Ding, Q., Huang, Y., Li, Y. X., Li, Y., Niu, Y. Y., and Wang, X. L.: Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9, Reprod. Fertil. Dev., 30, 1616–1621, https://doi.org/10.1071/rd18086, 2018.
Zuo, B. Y., Qian, H. G., Wang, Z. Y., Wang, X., Nisa, N., Bayier, A., Ying, S. J., Hu, X. L., Gong, C. H., Guo, Z. Q., and Wang, F.: A study on BMPR-IB genes of Bayanbulak sheep, Asian-Australas. J. Anim. Sci., 26, 36–42, https://doi.org/10.5713/ajas.2012.12238, 2013.