the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Polymorphism and expression of GLUD1 in relation to reproductive performance in Jining Grey goats
Wei Wang
Yongjuan Wang
Yufang Liu
Guiling Cao
Ran Di
Jinyu Wang
Mingxing Chu
Understanding the molecular mechanism of mammalian reproduction (puberty and prolificacy) will play a part in improving animal reproductive performance. GLUD1 (glutamate dehydrogenase 1) is important for mammalian reproduction, as shown in previous studies; however, its roles in puberty and prolificacy have rarely been reported. In this study, we designed seven pairs of primers (P1 to P7) for cloning and sequencing genomic DNA of Jining Grey goats and Liaoning Cashmere goats. Primer 8 (P8) was designed to detect single nucleotide polymorphism (SNP) of the GLUD1 in both sexually precocious and high-fecundity breeds (Jining Grey, Nanjiang Brown and Matou goats) and sexually late-maturing and low-fecundity breeds (Liaoning Cashmere, Inner Mongolia Cashmere and Taihang goats) by PCR-RFLP (restriction fragment length polymorphism). The real-time quantitative polymerase chain reaction (RT-qPCR) technique was used to detect the expression of GLUD1 in a variety of tissues. The results showed that the A197C mutation was only found in the amplification product of P6. For this SNP locus, only two genotypes (AA and AC) were detected in Nanjiang Brown goats, while three genotypes (AA, AC and CC) were detected in the other five breeds. In Jining Grey goats, the frequency of genotypes AA, AC and CC was 0.69, 0.26 and 0.05, respectively. In Jining Grey goats, AA genotype had 0.54 (P<0.05) and 0.3 (P<0.05) more kids than the CC and AC genotype, respectively, and no significant difference (P>0.05) was found in kidding number between the AC and CC genotype. GLUD1 was expressed in five tissues of different developmental stages. The expression level of GLUD1 in the hypothalamus was higher than that in the other four tissues except during puberty of Liaoning Cashmere goats. In puberty in goats, GLUD1 expression was significantly higher in ovaries than that in the juvenile period (P<0.01). RT-qPCR results showed that the expression of GLUD1 in ovaries may relate to the puberty of goats. The present study preliminarily indicated that there might be an association between the 197 locus of GLUD1 and sexual precocity in goats, and allele A of GLUD1 was a potential DNA marker for improving kidding number in Jining Grey goats.
- Article
(610 KB) - Full-text XML
-
Supplement
(566 KB) - BibTeX
- EndNote
Glutamate is the dominant excitatory amino acid neurotransmitter in neuroendocrine regulation. The availability of releasable glutamate from neurochemicals promotes a rapid increase in gonadotropin-releasing hormone (GnRH) secretion and enhanced GnRH mRNA and protein content, which was essential for controlling the onset of puberty (Van Den Pol et al., 1990; Brann and Mahesh, 1997; Terasawa and Fernandez, 2001). Previous studies revealed that the expression levels of glutamate were extremely low in pre-puberty; increased markedly in early puberty; and then maintained higher levels in puberty, which were associated with the expression of hypothalamic glutamate dehydrogenase (GDH) (Erecińska and Silver, 1990; Bourguignon et al., 1995; Terasawa et al., 1999; Roth et al., 2006). Significant alteration in the glutamate dehydrogenase activities may decrease the metabolism of glutamate, leading to the accumulation of extracellular glutamate (Malthankar-Phatak et al., 2006). Both glutamate and GDH were predominantly expressed in glial cells, suggesting that glial-cell-derived GDH output played a major role in the regulation of GnRH release in puberty (Terasawa and Fernandez, 2001). GLUD1 (glutamate dehydrogenase 1) is one of the enzymes which acts in the process of ammonia assimilation and is a hexameric enzyme with wide distribution. Some studies indicated that hormonal changes at puberty always had an extremely large impact on seizures and epilepsy (Mmako and Magazi, 2017; Odintsova et al., 2017; van Luijtelaar et al., 2017; Stefanidou and Montouris, 2020). Epilepsy onset was prevalent at the stages of the beginning of estradiol level increase and its ovulatory peaks (Odintsova et al., 2017; van Luijtelaar et al., 2017), suggesting that GLUD1 might play a role in precocious puberty.
Regulated by multiple endogenous, genetic and environmental factors, puberty is a complex and dynamic period in the development of goats, which is characterized by sexual maturation and the onset of reproductive capability. Puberty is controlled by glial cells, which regulate the availability of glutamate to the neuronal-glial network controlling the gonadotropin-releasing hormone (GnRH) (Roth et al., 2006). Puberty is the period of transition between infertility and fertility. Speeding up puberty transition would greatly reduce the production cost of reserve goats. In humans, shortened puberty is a sign of precocious puberty, which is mostly pathological (Sultan et al., 2018). However, some animal species have evolved precocious traits that can be stably inherited. Therefore, early sexual maturity may be an important quantitative trait in animal production. Genes associated with precocious puberty have attracted the attention of many scientists; ongoing research is likely to find out the main role of genes or closely linked molecular markers, effectively used to provide strong theoretical support for the prevention of human sexual premature disease and the breeding of animal varieties of early maturity and high fecundity (Maitra et al., 2014). During late postnatal development, GnRH in the hypothalamus drives the synthesis and secretion of follicle-stimulating hormone (FSH) and luteinizing hormone (LH), which are secreted through the hypothalamic–pituitary–ovarian axis and stimulate the gonads to trigger puberty. The secretion of GnRH neurons depended on various factors, such as γ-aminobutyric acid (GABA), G-protein-coupled receptor 54 (GPR54) and glutamate (Iremonger et al., 2010; Pineda et al., 2010; Watanabe et al., 2014). The kidding number is one of the most important economic traits in goats. Research on key genes of kidding can provide useful information for marker-assisted breeding to improve kidding number. Previous studies have proved that gene polymorphism can affect the reproductive performance of goat (Maitra et al., 2014; Tao et al., 2020).
Precocious puberty refers to the premature appearance of secondary sexual characteristics in individual goats, while delayed puberty is the opposite. Nanjiang Brown goats, Matou goats and Jining Grey goats are highly prolific local breeds in PR China. Taihang goats, Inner Mongolia Cashmere goats and Liaoning Cashmere goats are lowly prolific local breeds in PR China. Jining Grey goats are precocious and generally reach estrus at the age of 2–3 months (Tu, 1989); Nanjiang Brown goats are precocious and begin estrus at the age of 4 months (Du, 2011); Matou goats mature earlier, and the average age of estrus of goats is 107.9 d (Tu, 1989); Liaoning Cashmere goats mature late and reach estrus at the age of 5–6 months (Tu, 1989); Inner Mongolia Cashmere goats mature late and reach estrus at the age of 5–6 months (Tu, 1989); and Taihang goats generally reach estrus at 6–7 months of age (Tu, 1989).
In this study, GLUD1 single nucleotide polymorphisms (SNPs) were analyzed in early-puberty goats (Nanjiang Brown goats, Matou goats and Jining Grey goats) and delayed-puberty goat breeds (Taihang goats, Inner Mongolia Cashmere goats and Liaoning Cashmere goats) by sequencing and restriction fragment length polymorphism (RFLP). The real-time quantitative polymerase chain reaction (RT-qPCR) technique was used to study the expression of GLUD1 in different tissues of the hypothalamus, pituitary gland, ovaries, uterus and adrenal gland of Jining Grey goats and Liaoning Cashmere goats at 30 ± 5, 90 ± 5 and 160–180 d. The present study aimed to find genetic markers related to sexual precocity and high fertility in goats and to provide a scientific basis for marker-assisted early selection of reproductive performance.
2.1 Animal preparation and sample collection
Jugular blood samples from 475 does (2–3 years) were processed with phenol–chloroform and dissolved in ddH2O. Among the six goat breeds involved in the present study, there were three precocious-puberty goat breeds (Nanjiang Brown goats, n=30; Matou goats, n=48; Jining Grey goats, n=212) and three delayed-puberty goat breeds (Taihang goats, n=55; Inner Mongolia Cashmere goats, n=59; Liaoning Cashmere goats, n=71) (Table 1, Supplement Fig. S1) (Du, 2011; Feng et al., 2011). Five tissue samples including the hypothalamus, pituitary gland, ovaries, uterus and adrenal gland were taken from Jining Grey goats and Liaoning Cashmere goats. All the tissue samples were collected within 1 h after the goats were slaughtered and preserved in a 2 mL RNase-Free frozen storage tube in liquid nitrogen. Then, all samples were taken back to the laboratory and stored at −80∘C.
The 212 Jining Grey goats were randomly selected as the offspring of five bucks, whose parities, kidding months and litter size were recorded. The first parity (n=62), the second parity (n=68) and the third parity (n=82) of Jining Grey goats were defined as three parities. According to the actual kidding time, March to May was taken as spring kidding (n=56), June to August as summer kidding (n=47), September to November as autumn kidding (n=65) and December to February as winter kidding (n=44).
2.2 RNA extraction and primer design
Total RNA was isolated from samples using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA purity was confirmed with a NanoPhotometer spectrophotometer (IMPLEN, CA, USA), and concentration and integrity were assessed using electrophoresis and the RNA 6000 Nano assay kit of the 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA).
To amplify the GLUD1 coding sequence of goats, eight primer pairs were designed according to DNA fragments (contig) containing the region of interest using Primer Premier software (version 5.0) and Oligo software (version 6.0), and synthesized by Sangon Biotech Co. Ltd. (Shanghai, China) (Table 2).
2.3 PCR amplification and real-time quantitative PCR
The PCR amplification reaction system is shown in Table 3. The reaction program is as follows: 95∘C predenaturation for 5 min, 95∘C denaturation for 30 s, annealing for 30 s and 72∘C extension for 30 s for 35 cycles and then 72 ∘C extension for 5 min. After PCR, the product was stored at 4∘C.
The experimental process of RT-qPCR was the same as described by Hu et al. (2020) in our laboratory. In short, cDNA was synthesized according to the steps provided by the reverse transcription kit; 2 × Taq PCR Master Mix was used to perform PCR on GLUD1, and SYBR®Premix Ex Taq™ II was finally used for RT-qPCR. The experiment was carried out using the Roche Light Cycler®480 II fluorescent quantitative PCR instrument, with GAPDH as the internal reference gene, and this was repeated for each sample three times. The 2−ΔΔct method (Hu et al., 2021) was used to calculate the relative expression of the target gene. SPSS 24.0 statistical software was used to perform a one-way analysis of variance (ANOVA) on the data, and the least significant difference (LSD) test was used to evaluate differences between groups. The P<0.05 value was considered to indicate significance.
2.4 Cloning and sequencing
Following the instructions of the manufacturer, the amplified fragments were separated on agarose gels (1.5 %) and purified with a DNA fragment quick recover kit (TaKaRa, Dalian, China), and they were then inserted into the pMD18-T vector (TaKaRa, Dalian, China). The recombinant plasmids were transformed into E.coli JMl09 competent cells. Positive clones were identified by restriction enzyme digestion and sequenced by Beijing Genomics Institute (China). To identify different nucleotides, DNAMAN (v. 5.2.2) was used for sequence analysis.
2.5 PCR-RFLP analysis
The restriction enzyme digestion mixture contained 1 µL 10 × NEB buffer, 0.3 µL restriction endonuclease BsoBI (10 U µL−1, NEB), 4 µL PCR product (P8 primer) and 4.7 µL ddH2O. The reaction was held at 37∘C for 5 h. The digested products were analyzed by electrophoresis on 12 % polyacrylamide gel (acrylamide : bis-acrylamide = 39:1). AlphaImager™ 2200 and 1220 Documentation and Analysis Systems (Alpha Innotech Corporation, San Leandro, CA, USA) were used to analyze and count genotypes.
2.6 Statistical analyses
Allele frequencies, genotype frequencies, p values, polymorphism information content (PIC), heterozygosity (HE) and the number of effective alleles (NE) were calculated using the data obtained from genotyping results. The chi-squared test was used to detect the Hardy–Weinberg (H-W) equilibrium of each mutation site in six goat populations (Ortega et al., 2016). We performed least-squares analysis of variance by SAS with the following model to analyze the genotypes of primer P8 and compare the differences in litter size of Jining Grey goats with different genotypes.
where yijklm is the recorded value of litter size, μ is the overall population mean, Si is the fixed effect of the male goat (i=1, 2, 3, 4, 5), KSj is the fixed effect of the kidding season (j=1, 2, 3, 4), Pk is the fixed effect of the parity (k=1, 2, 3), Gl is the fixed effect of the genotype and eijklm is a random residual effect.
3.1 PCR amplification
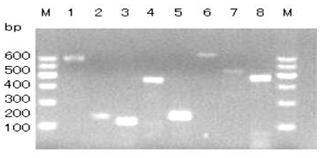
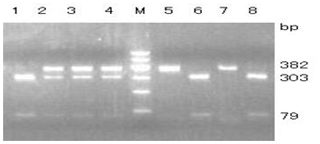
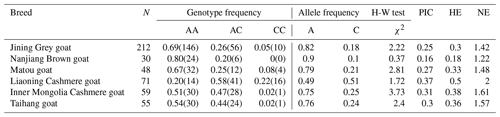
The GLUD1 of six goat breeds was amplified by primers P1 to P8, and the PCR products were detected by 1.5 % agarose gel electrophoresis. The results showed that the amplified fragments were consistent with the expected size, and specific fragments were obtained (Fig. 1).
3.2 Sequence analysis of amplification fragments
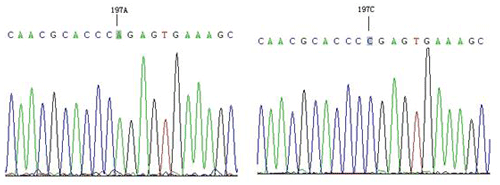
The results showed that only one mutation site (A197C) was found in the amplified product of P6, and fragment P6 corresponds to exon8–exon9. Compared with the AA genotype, the CC genotype had an A → C mutation at position 197 (Fig. 2), which did not cause amino acid changes and was a silent mutation.
3.3 RFLP analysis of the amplified fragment
One pair of new primers P8 was designed to detect the A → C mutation at position 197. The restriction enzyme BsoBI was used to directly perform RFLP detection. As shown in Fig. 3, there were three genotypes: AA (382 bp), AC (79/303/382 bp) and CC (79/303 bp).
3.4 The expression level of goat GLUD1 in various tissues
We calculated the RT-qPCR results of the target gene of each sample. We also contrasted the expression difference in mRNA transcription level of the target gene in other samples relative to the reference sample (the average value of the ovaries in the juvenile period of Jining Grey goats was the reference sample). The results are shown in Table 4.
Table 4The expression of GLUD1 in different tissues of Jining Grey and Liaoning Cashmere goats.
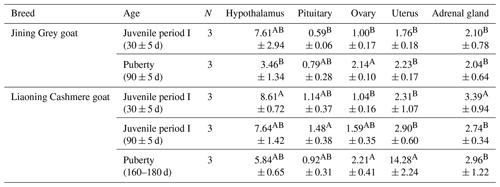
The values are averages. In the same column, different uppercase letters next to means indicate differences (P<0.01), and the same letters indicate nonsignificant differences. In the same row, different uppercase letters next to means indicate differences (P<0.01), and the same letters indicate nonsignificant differences. N: number of individual goats.
GLUD1 was expressed in five tissues at each developmental stage of Jining Grey goats and Liaoning Cashmere goats (Table 4). The results showed that the expression of GLUD1 in the hypothalamus of high-fecundity breeds (Jining Grey goats) was significantly higher than that in the other four tissues (P<0.01), and it was the lowest in the pituitary. The expression level of GLUD1 in the hypothalamus in the juvenile period I of Jining Grey goats was significantly higher than that in puberty (P<0.01). There were no significant differences between the expression levels of GLUD1 in the other three tissues of the Jining Grey goats in different periods. In the low-fecundity breed (Liaoning Cashmere goats), the expression of GLUD1 in the hypothalamus in the juvenile period (I and II) was significantly higher than that in the other four tissues (P<0.01). However, the expression of GLUD1 in the hypothalamus and uterus at puberty was significantly higher than that of the other three tissues (P<0.01). Except for the juvenile stage I of Liaoning Cashmere goats, the expression of GLUD1 was the lowest in the pituitary at all stages (P<0.01).
In juvenile period I, the expression level of GLUD1 in the hypothalamus of Liaoning Cashmere goats was significantly higher than that in Jining Grey goats (P<0.01). During puberty, the expression levels of GLUD1 in the uterus of Liaoning Cashmere goats were lower than those in Jining Grey goats (P<0.01).
3.5 Allele and genotype frequencies of GLUD1 in six goat breeds
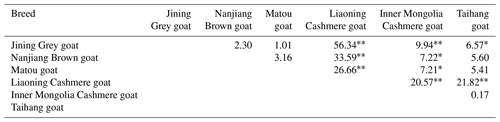
The allele and genotype frequencies of the GLUD1 mutation site in six goat breeds were shown in Table 5. The results showed that the A allele was the major allele in the five goat breeds, while only the C allele in the Liaoning Cashmere goat was the dominant allele. All populations were in a Hardy–Weinberg equilibrium state (P>0.05).
3.6 Genotype distribution of GLUD1 in six goat breeds
The differences in the distribution of different genotypes at the A197C locus among the six goat breeds were tested (Table 6). The results showed that there was a significant difference in the distribution of genotypes at this locus between the high-fecundity breed (Jining Grey goats) and the low-fecundity breeds (Liaoning Cashmere goats, Inner Mongolia Cashmere goats and Taihang goats) (P<0.01). Besides, another significant difference was found between the other high-fecundity breeds (Nanjiang Brown goats and Matou goats) and the low-fecundity breed (Liaoning Cashmere goats) (P<0.01). There was also a significant difference between the high-fecundity breeds (Nanjiang Brown goats and Matou goats) and the low-fecundity breed (Inner Mongolia Cashmere goats) (P<0.05). These results indicated that this locus might be associated with the litter size of goats.
3.7 The effect of GLUD1 on litter size of Jining Grey goats
The average litter size and standard errors of different genotypes of GLUD1 in Jining Grey goats are shown in Table 7. For the 197 locus, the average litter size of AA type Jining Grey goats had 0.54 (P<0.05) and 0.30 (P<0.05) kids more than that of CC- and AC-type Jining Grey goats, respectively. There was no significant difference in litter size between AC and CC types.
Table 7Least-squares mean and standard error for litter size of different genotypes of the 197 locus of GLUD1 in Jining Grey goats.

Least-squares means with the same superscript have no significant difference (P>0.05). The least-squares means with the different superscripts differ significantly (P<0.05).
4.1 Expression of GLUD1
In recent studies, GLUD1 mainly functioned as an amino-acid-metabolism-related gene, and cytoplasmic GLUD1 degradation was shown to limit protein synthesis (Shao et al., 2021). In Saccharomyces cerevisiae, glutamate can be synthesized from α-ketoglutarate and ammonium through the action of the NADP-dependent glutamate dehydrogenases GLUD1 and GDH3 (Mara et al., 2018). However, the role played by the expression of glutamate dehydrogenase in pubertal development cannot be ignored. It has been found that the expression of glutamate dehydrogenase in the hypothalamus of female rats increased during the development of puberty, while the expression of glutamate synthetase catalyzed the metabolism of glutamate from glutamine (Hasan Mahmood et al., 2018; Ibrahim et al., 2020; Uddin et al., 2020). Spanaki et al. (2021) mentioned that GLUD1 was intensively expressed in estrogen-producing cells in human ovaries and placenta, and it was a housekeeping enzyme (Spanaki et al., 2017), which was also reflected in our study. The results of our study showed that GLUD1 was highly expressed in ovaries of Jining Grey goats and Liaoning Cashmere goats at various developmental stages, and the expression in the first estrus was higher than that in juvenile period. It was speculated that the change of GLUD1 expression in ovaries affected the first estrus. In human steroidogenic organs, GLUD1 was highly expressed in steroid synthesizing cells with this expression pattern, matching that of the cholesterol side-chain system involved in steroid biosynthesis. However, there were few studies focused on the expression of GLUD1 in goats. The expression of GLUD1 in five tissues of goats was analyzed in this study, and there was no significant difference in the expression of GLUD1 in the hypothalamus between Jining Grey goats and Liaoning Cashmere goats in the same period. Except for Liaoning Cashmere goats at the first stage of estrus, the expression levels in the hypothalamus were shown to be higher than those in the other four tissues in all other stages.
4.2 Polymorphism and function of GLUD1
In this study, goat GLUD1 had a mutation site of A197C in the exon 8 region, which did not alter the amino acid sequence and was a silent mutation. This study also showed that there were obvious differences between sexually early-maturing breeds and late-maturing breeds in the 197 mutation site of different genotypes. Therefore, it was inferred that A197C of GLUD1 may have an association with the precocious puberty of goats. At present, plenty of studies have focused on the litter size in livestock (Tao et al., 2020, 2021). Previous studies showed that polymorphisms of BMPR1B could affect the sheep litter size and that BMPR1B affects ovulation in mammals by promoting follicular development and ovarian granulosa cell proliferation (Hu et al., 2020; Wen et al., 2021). However, few studies associating the GLUD1 polymorphisms with litter size have been published. It was proved that gene polymorphism can affect the number of kidding, which was consistent with previous studies. In previous studies, GLUD1 was found to contribute to glucose-stimulated insulin secretion in mouse beta cells (Lin et al., 2012; Goldsmith et al., 2014), indicating that its regulation of autophagy was sensitive to the metabolic environment. GLUD1 could reduce NAD+ and/or NADP+ to NADH and/or NADPH, which promoted biological processes such as ammonia handling and Krebs cycle anaplerosis by linking amino acids to carbohydrate metabolism (Spanaki et al., 2017). Moreover, mammalian GDH (hGLUD1 in humans) has acquired an energy-sensing mechanism (GTP–ADP binding) that regulated enzyme function according to cellular energy needs (Mastorodemos et al., 2005). However, the mechanism of GLUD1 in goat reproduction has not been clarified. Our study preliminarily indicated that allele A at locus 197 of the GLUD1 might increase the kidding number in Jining Grey goats, which might be a potentially effective DNA marker for increasing kidding of goats. The samples of Jining Grey goats in this study were all from the same breeding farm, so there might be some kinship between individuals, and the effective population size of the tested goats was reduced, which affected the degree of association between GLUD1 and kidding number. This also confirms the previous findings that genetic polymorphisms can affect kidding numbers in goats and that studying the role of GLUD1 polymorphisms can provide useful DNA markers for selecting quality individuals by marker-assisted selection (MAS) and contribute to goat breeding (Yang et al., 2021). In the next step, we will increase the number of goat breeds, expand the number of samples within the breed and conduct further studies.
All the experimental procedures were authorized by the Science Research Department of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (IAS-CAAS; Beijing, China). The study has also been ethically approved by the Animal Ethics Committee of the IAS (IAS2020-62).
In this study, it was found that GLUD1 was expressed in five tissues at all developmental stages in goats. Except for the puberty of Liaoning Cashmere goat, the expression in the hypothalamus at other stages was higher than that in the other four tissues. In ovaries, the expression of GLUD1 in puberty was significantly higher than that in infancy, which speculated that the change of GLUD1 expression in ovaries affected puberty. The difference test of our research also showed that GLUD1 might affect puberty of goats. A mutation at position 197 of GLUD1 has been detected, and association analysis showed that allele A of this locus is associated with an increase in the number of kidding in Jining Grey goats.
No data sets were used in this article.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-66-411-2023-supplement.
Conceptualization, WW and YW; methodology, WW; software, YL and GC; writing (original draft preparation), WW; writing (review and editing), JW; supervision, MC; project administration, MC; funding acquisition, MC. All authors have read and agreed to the published version of the manuscript.
The contact author has declared that none of the authors has any competing interests.
The Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS; Beijing, P.R. China), has approved all procedures involving laboratory animals. Ethics approval was also granted by the Animal Ethics Committee of IAS-CAAS (no. IAS2020-63; 27 April 2020).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This work was funded by the National Natural Science Foundation of China (32102509), China Agriculture Research System of MOF and MARA (CARS-38), and Agricultural Science and Technology Innovation Program of China (CAAS-ZDRW202106 and ASTIP-IAS13).
This research has been supported by the National Natural Science Foundation of China (grant no. 32102509), the Ministry of Finance (grant no. CARS-38), and the Agricultural Science and Technology Innovation Program (grant nos. CAAS-ZDRW202106 and ASTIP-IAS13).
This paper was edited by Henry Reyer and reviewed by Samia Kdidi, Shahin Eghbalsaied, and three anonymous referees.
Bourguignon, J. P., Gerard, A., Alvarez, Gonzalez, M. L., Purnelle, G., and Franchimont, P.: Endogenous glutamate involvement in pulsatile secretion of gonadotropin-releasing hormone: evidence from effect of glutamine and developmental changes, Endocrinology, 136, 9110–9116, https://doi.org/10.1210/endo.136.3.7867599, 1995.
Brann, D. W. and Mahesh, V. B.: Excitatory amino acids: evidence for a role in the control of reproduction and anterior pituitary hormone secretion, Endocr. Rev., 18, 678–700, https://doi.org/10.1210/edrv.18.5.0311, 1997.
Du, L. X.: Animal Genetic Resources in China-Sheep and Goats, China Agriculture Press, Beijing, China, 238–241, 242–244, 251–254, 283–286, 303–305, 423–425, 2011.
Erecińska, M. and Silver, I. A.: Metabolism and role of glutamate in mammalian brain, Prog. Neurobiol., 35, 245–296, https://doi.org/10.1016/0301-0082(90)90013-7, 1990.
Feng, T., Geng, C. X., Lang, X. Z., Chu, M. X., Cao, G. L., Di, R., Fang, L., Chen, H. Q., Liu, X. L., and Li, N.: Polymorphisms of caprine GDF9 gene and their association with litter size in Jining Grey goats, Mol. Biol. Rep., 38, 5189–5197, https://https://doi.org/10.1007/s11033-010-0669-y, 2011.
Goldsmith, J., Levine, B., and Debnath, J.: Autophagy and cancer metabolism, Methods Enzymol., 542, 25–57, https://doi.org/10.1016/B978-0-12-416618-9.00002-9, 2014.
Hasan Mahmood, A. S. M., Uddin, M. M., Ibrahim, M. M. H., Mandal, S. K., Alhamami, H. N., and Briski, K. P.: Sex differences in forebrain estrogen receptor regulation of hypoglycemic patterns of counter-regulatory hormone secretion and ventromedial hypothalamic nucleus glucoregulatory neurotransmitter and astrocyte glycogen metabolic enzyme expression, Neuropeptides, 72, 65–74, https://doi.org/10.1016/j.npep.2018.10.003, 2018.
Hu, W., Tang, J., Zhang, Z., Tang, Q., Yan, Y., Wang, P., Wang, X., Liu, Q., Guo, X., Jin, M., Zhang, Y., Di, R., and Chu, M.: Polymorphisms in the ASMT and ADAMTS1 gene may increase litter size in goats, Vet. Med. Sci., 6, 775–787, https://doi.org/10.1002/vms3.301, 2020.
Hu, W. P., Liu, M. Q., Tian, Z. L., Liu, Q. Y., Zhang, Z. B., Tang, J. S., He, X. Y., Zhu, Y. Y., Wang, Y. Y., and Chu, M. X.: Polymorphism, expression and structure analysis of key genes in the ovarian steroidogenesis pathway in sheep (Ovis aries), Vet. Med. Sci., 7, 1303–1315, https://doi.org/10.1002/vms3.485, 2021.
Ibrahim, M. M. H., Uddin, M. M., Bheemanapally, K., and Briski, K. P.: Sex-dimorphic aromatase regulation of ventromedial hypothalamic nucleus glycogen content in euglycemic and insulin-induced hypoglycemic rats, Neurosci. Lett., 737, 135284, https://doi.org/10.1016/j.neulet.2020.135284, 2020.
Iremonger, K. J., Constantin, S., Liu, X., and Herbison, A. E.: Glutamate regulation of GnRH neuron excitability, Brain Res., 1364, 35–43, https://doi.org/10.1016/j.brainres.2010.08.071, 2010.
Lin, T. C., Chen, Y. R., Kensicki, E., Li, A. Y., Kong, M., Li, Y., Mohney, R. P., Shen, H. M., Stiles, B., Mizushima, N., Lin, L. I., and Ann, D. K.: Autophagy: resetting glutamine-dependent metabolism and oxygen consumption, Autophagy, 8, 1477–1493, https://doi.org/10.4161/auto.21228, 2012.
Maitra, A., Sharma, R., Ahlawat, S., Tantia, M. S., Roy, M., and Prakash, V.: Association analysis of polymorphisms in caprine Kiss1 gene with reproductive traits, Anim. Reprod. Sci., 151, 71–77, https://doi.org/10.1016/j.anireprosci.2014.09.013, 2014.
Malthankar-Phatak, G. H., De Lanerolle, N., Eid, T., Spencer, D. D., Behar, K. L., Spencer, S. S., Kim, J. H., and Lai, J. C.: Differential glutamate dehydrogenase (GDH) activity profile in patients with temporal lobe epilepsy, Epilepsia, 47, 1292–1299, https://doi.org/10.1111/j.1528-1167.2006.00543.x, 2006.
Mara, P., Fragiadakis, G. S., Gkountromichos, F., and Alexandraki, D.: The pleiotropic effects of the glutamate dehydrogenase (GDH) pathway in Saccharomyces cerevisiae, Microb. Cell Fact., 17, 170, https://doi.org/10.1186/s12934-018-1018-4, 2018.
Mastorodemos, V., Zaganas, I., Spanaki, C., Bessa, M., and Plaitakis, A.: Molecular basis of human glutamate dehydrogenase regulation under changing energy demands, J. Neurosci. Res., 79, 65–73, https://doi.org/10.1002/jnr.20353, 2005.
Mmako, M. and Magazi, D.: Women living with Epilepsy, a review and guide for the family physician, Mental Health Matters, 4, 18–20, 2017.
Odintsova, G., Chugunova, A., and Nesterova, S.: Epilepsy onset in puberty, Eur. J. Paediatr. Neuro., 21, e108, https://doi.org/10.1016/j.ejpn.2017.04.767, 2017.
Ortega, M., Denicol, A., Cole, J., Null, D., and Hansen, P.: Use of single nucleotide polymorphisms in candidate genes associated with daughter pregnancy rate for prediction of genetic merit for reproduction in Holstein cows, Anim. Genet., 47, 288–297, https://doi.org/10.1111/age.12420, 2016.
Pineda, R., Aguilar, E., Pinilla, L., and Tena-Sempere, M.: Physiological roles of the kisspeptin/GPR54 system in the neuroendocrine control of reproduction, Prog. Brain Res., 181, 55–77, https://doi.org/10.1016/S0079-6123(08)81005-9, 2010.
Roth, C. L., McCormack, A. L., Lomniczi, A., Mungenast, A. E., and Ojeda, S. R. J.: Quantitative proteomics identifies a change in glial glutamate metabolism at the time of female puberty, Mol. Cell Endocrinol., 254, 51–59, https://doi.org/10.1016/j.mce.2006.04.017, 2006.
Shao, J., Shi, T., Yu, H., Ding, Y., Li, L., Wang, X., and Wang, X.: Cytosolic GLUD1 degradation restricts protein synthesis to sustain tumor cell survival following amino acid deprivation, Embo J., 40, e107480, https://doi.org/10.15252/embj.2020107480, 2021.
Spanaki, C., Kotzamani, D., and Plaitakis, A.: Widening Spectrum of Cellular and Subcellular Expression of Human GLUD1 and GLUD2 Glutamate Dehydrogenases Suggests Novel Functions, Neurochem. Res., 42, 92–107, https://doi.org/10.1007/s11064-016-1986-x, 2017.
Stefanidou, M. and Montouris, G.: Reproductive and sexual health concerns in transition-age adolescents and young adults with epilepsy, Semin. Pediatr. Neurol., 36, 100855, https://doi.org/10.1016/j.spen.2020.100855, 2020.
Sultan, C., Gaspari, L., Maimoun, L., Kalfa, N., and Paris, F.: Disorders of puberty, Best practice & research, Clin. Obstetr. Gynaecol., 48, 62–89, https://doi.org/10.1016/j.bpobgyn.2017.11.004, 2018.
Tao, L., He, X. Y., Jiang, Y. T., Lan, R., Li, M., Li, Z. M., Yang, W. F., Hong, Q. H., and Chu, M. X.: Combined approaches to reveal genes associated with litter size in Yunshang black goats, Anim. Genet., 51, 924–934, https://doi.org/10.1111/age.12999, 2020.
Tao, L., He, X. Y., Wang, F. Y., Pan, L. X., Wang, X. Y., Gan, S. Q., Di, R., and Chu, M. X.: Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep, Anim. Genet., 52, 545–549, https://doi.org/10.1111/age.13078, 2021.
Terasawa, E. and Fernandez, D. L.: Neurobiological mechanisms of the onset of puberty in primates, Endocr. Rev., 22, 111–151, https://doi.org/10.1210/edrv.22.1.0418, 2001.
Terasawa, E., Luchansky, L., Kasuya, E., and Nyberg, C.: An increase in glutamate release follows a decrease in gamma aminobutyric acid and the pubertal increase in luteinizing hormone releasing hormone release in the female rhesus monkeys, J. Neuroendocrinol., 11, 275–282, https://doi.org/10.1046/j.1365-2826.1999.00325.x, 1999.
Tu, Y. R.: The sheep and goat breeds in China, Shanghai Science and Technology Press, Shanghai, 98–101, 104–106, 92–93, 88–90, 93–95, 1989.
Uddin, M. M., Ibrahim, M. M. H., and Briski, K. P.: Sex-dimorphic neuroestradiol regulation of ventromedial hypothalamic nucleus glucoregulatory transmitter and glycogen metabolism enzyme protein expression in the rat, BMC Neurosci., 21, 51, https://doi.org/10.1186/s12868-020-00598-w, 2020.
Van Den Pol, A. N., Wuarin, J. P., and Dudek, F. E.: Glutamate, the dominant excitatory transmitter in neuroendocrine regulation, Science, 250, 1276–1278, https://doi.org/10.1126/science.1978759, 1990.
van Luijtelaar, E., Tolmacheva, E., and Budziszewska, B.: Hormones and absence epilepsy, Neurosci. Biobehav. Psychol., 1, 488–495, https://doi.org/10.1016/B978-0-12-809324-5.00083-3, 2017.
Watanabe, M., Fukuda, A., and Nabekura, J.: The role of GABA in the regulation of GnRH neurons, Front. Neurosci., 8, 387, https://doi.org/10.3389/fnins.2014.00387, 2014.
Wen, Y.-L., Guo, X.-F., Ma, L., Zhang, X.-S., Zhang, J.-L., Zhao, S.-G., and Chu, M.-X.: The expression and mutation of BMPR1B and its association with litter size in small-tail Han sheep (Ovis aries), Arch. Anim. Breed., 64, 211–221, https://doi.org/10.5194/aab-64-211-2021, 2021.
Yang, Y., Sun, X., Cui, W., Liu, N., Wang, K., Qu, L., and Pan, C. The detection of mutation within goat cell division cycle 25 A and its effect on kidding number, Anim. Biotechnol., 3, 1504–1509, https://doi.org/10.1080/10495398.2021.1910519, 2021.