the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Antibiotic resistance and biofilm formation in Staphylococcus aureus isolated from dairy cows at the stage of subclinical mastitis in northern Kazakhstan
Raushan Rychshanova
Anara Mendybayeva
Bartosz Miciński
Nurlan Mamiyev
Pavel Shevchenko
Zhanaidar Bermukhametov
Bartosz Orzechowski
Jan Miciński
Staphylococcus aureus is an important causative agent of subclinical bovine mastitis worldwide. The aim of this research was to study the ability of S. aureus to form biofilms. Additionally, we examined the genes involved in cell resistance and sensitivity to antibiotics. Samples were collected from December 2020 to May 2021 from Simmental and black-and-white cows. The study was carried out on a total number of 643 cows, of which 278 (23 %) were in the subclinical mastitis stage. Finally, 64 S. aureus isolates were isolated and identified. The highest level of phenotypic resistance was observed to antibiotics of the tetracycline (tetracycline – 48.4 %, doxycycline – 32.8 %) and β-lactam (ampicillin – 45.3 %, penicillin – 45.3 %) groups. The genes encoding antibiotic resistance were characterized with the polymerase chain reaction method: blaZ in 30 isolates, mecA in 1 isolate, ermC in 15 isolates, aph (3) in 2 isolates, tetK in 19 isolates, tetM in 9 isolates. The tested S. aureus isolates had the ability to form biofilms in 76.6 % () of cases. Of these, 69.4 % were resistant to at least one antibiotic. The obtained results have shown that S. aureus, identified in cows with subclinical mastitis, was resistant mainly to tetracycline and β-lactam antibiotics. In addition, S. aureus isolates expressed resistance genes to the above drugs and had the ability to form biofilm. This study will help to identify the extent of antibiotic resistance and monitor S. aureus contamination of raw milk.
- Article
(651 KB) - Full-text XML
- BibTeX
- EndNote
Mastitis is one of the global problems of the dairy industry. The problem with treating mastitis is that there is currently no clear-cut cause of the disease. Staphylococci, streptococci, and E. coli have been shown to be of particular importance to the etiology of this disease (Shathele, 2009). According to Wang et al. (2018), staphylococcal infection is one of the main causes of mastitis in dairy cows, with relatively low cure rates. The microorganisms have the ability to form biofilms and release toxins that cause the formation of a tissue barrier by the host, as well as the spread of new strains with antibiotic-resistant properties (Timakov et al., 2015; Thiran et al., 2018). This is the cause of large financial and economic losses for cattle breeders (Aslantaş and Demir, 2016). Staphylococcus aureus ranks third among the most common foodborne pathogens in the world (Hennekinne et al., 2012). It has been shown that milk and dairy products are the main potential sources of S. aureus infections in humans (Ali Gharieb et al., 2020).
It is well known that levels of antimicrobial resistance vary significantly, not only between countries but also between regions and farms. For example, estimates of the prevalence of penicillin resistance in S. aureus isolated from cattle in the United States range from 30 % to 70 % (Barkema et al., 2006).
The aim of the research was to determine the proportion of subclinical cases in which S. aureus develops the ability to create biofilms and to examine genes responsible for the level of resistance and sensitivity to antibiotics.
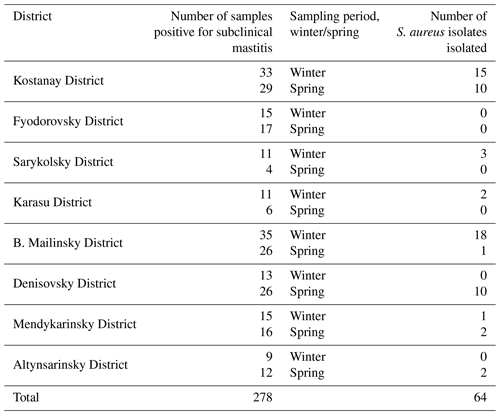
Between December 2020 and May 2021, all milk samples were taken from cows of black-and-white Holsteinized (n=515) and Simmental (n=128) breeds aged from 2 to 9 years. The cattle used in the experiment were bred in a free-standing manner and milked with the use of a milking machine. A total number of 643 cow's milk samples were collected, of which 278 were at the stage of subclinical mastitis. Samples were taken from cows of dairy farms located in the northern Kostanay region (Kazakhstan). Out of 16 districts of the Kostanay region, samples were taken from 8 districts (Kostanay District, Fyodorovsky District, Sarykolsky District, Karasu District, B. Mailinsky District, Denisovsky District, Mendykarinsky District, Altynsarinsky District), in the territory of which the largest dairy farms of the region operate. The sampling season was winter (December, January, February) and spring (March, April, May).
2.1 Sample collection
The stage of subclinical mastitis (subclinical mastitis – clinical signs of mastitis are absent or mild) was determined by measuring the electrical resistance of milk with a Draminski 4×4 Q Mast apparatus (Poland). The absence of obvious clinical signs and a difference of more than 40–50 units between the maximum and minimum results for quarters in the examined cows indicated the beginning of subclinical mastitis. The first portions of milk were collected under aseptic conditions according to Oliver et al. (2004). The samples were transported to the laboratory immediately after collection.
2.2 Staphylococcal isolation
Isolation and identification of S. aureus were carried out according to State Standard (2016) with minor modifications. Briefly, at the first stage, a sample of milk was inoculated into salt broth in a ratio of 1:10, followed by incubation at 37±1 ∘C for 18–24 h. After incubation, inoculation was made on mannitol saline agar (HiMedia Laboratories, India) and incubated for 24 h at 37±1 ∘C. Typical colonies, flat or convex discs of yellow, lemon colour with smooth edges, were seeded onto the surface of meat-peptone agar (BIOKOMPAS-S, Russia). Haemolytic properties were determined by blood agar (HiMedia Laboratories, India). To confirm whether the isolates belonged to coagulase-positive staphylococci, Gram staining, tests for catalase (hydrogen peroxide, Kazakhstan) and coagulase (Microgen, Russia) were performed. To determine the belonging of the identified coagulase-positive staphylococci to S. aureus, the ability to ferment mannitol under anaerobic conditions was tested. The presence of deoxyribonuclease activity was investigated on a DNase medium (HiMedia Laboratories, India) with toluidine blue (HiMedia Laboratories, India).
Bacteria belonging to S. aureus were confirmed using the STAPHYtest 24 identification system.
2.3 Detection of antibiotic resistance
S. aureus isolates were tested for antibiotic susceptibility using the disc diffusion method (EUCAST Disk Diffusion Method for Antimicrobial Susceptibility Testing – Version 9.0 (January 2021)) on a Mueller–Hinton medium (HiMedia Laboratories, India). The following discs (Pasteur Institute of Epidemiology and Microbiology, Russia) were used for testing: ampicillin (10 µg), amoxicillin (25 µg), benzylpenicillin (10 U), cefoperazone (75 µg), cefoxitin (30 µg), streptomycin (10 µg), kanamycin (30 µg), neomycin (30 µg), gentamicin (120 µg), tetracycline (30 µg), doxycycline (30 µg), sulfamethoxazole with trimethoprim (), erythromycin (15 µg), tylosin (15 µg). Strain RCM-0056 was used as a positive control. The data were interpreted according to the recommendations taking into account EUCAST 2021 (EUCAST, 2021), CLSI 2019 (CLSI, 2019), and MI 2004 (MI 4.2.1890-04, 2004).
2.4 Biofilm formation
A biofilm formation test was performed on a 96-well polystyrene plate by crystal violet staining (Okulich, 2017). First, the studied cultures were incubated at 37±1 ∘C in tryptone-soy broth (HiMedia Laboratories, India) for 24 h, and the suspension concentration was adjusted to 0.5 McFarland. A total of 200 µL of the bacterial suspension was added to the wells of a polystyrene plate and incubated at 37±1 ∘C for 24 h. Pure broth was used as a negative control. After incubation, the medium with planktonic cells was removed and washed four times with distilled water. Then the biofilms were fixed on the plate by adding 160 µL of 2.5 % glutaraldehyde solution and kept for 5 min. Next, it was washed again with distilled water and stained with 0.25 % crystal violet (HiMedia Laboratories, India) solution for 5 min. The plate was washed and dried at 25 ∘C for 10 min. To extract the dye solution from the cells, 200 µL of 33 % glacial acetic acid solution was added to the wells. The optical density was measured on a spectrophotometer (Multiskan FC, Thermo Fisher, USA) at a wavelength of 620 nm (Okulich, 2017). Interpretation was carried out according to Aslantaş and Demir (2016), using a three-point scale of optical density: <0.120 (–), 0.120–0.240 (+), .
2.5 Determination of resistance genes by PCR
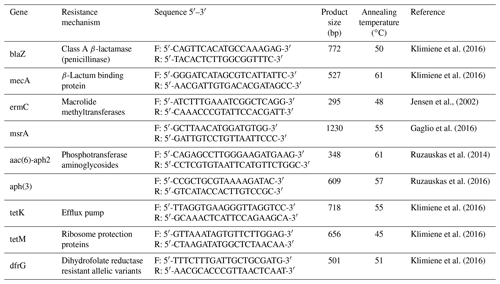
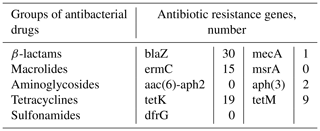
To determine genes related to antibacterial drugs, the polymerase chain reaction (PCR) method was used with the following primers: a group of β-lactams (blaZ, mecA, msrA), macrolides (ermC), aminoglycosides (aac(6)-aph2, aph(3)), tetracyclines (tetK, tetM), and sulfonamides (dfrG; Syntol, Russia; Table 1).
Bacterial DNA was isolated according to Chuzhebaeva et al. (2020) with the use of a thermal cycler (Applied Biosystems, Thermo Fisher Scientific, USA). The reaction mixture consisted of DreamTaq Green Master Mix, water, primers at a concentration of 10 pmol mL−1, and tested DNA. The PCR conditions were as follows: pre-denaturation at 94 ∘C for 5 min, next 30 cycles at 94 ∘C for 30 s, annealing at a temperature according to Table 1 for each primer used for 1 min, and elongation at 72 ∘C for 1 min. In the end, the final elongation at 72 ∘C for 10 min was performed. Visualization of the PCR product was performed on a 1.5 % agarose gel (Thermo Fisher Scientific, USA) with a QUANTUM UV transilluminator (1100SUPER-BRIGHT; Peqlab, Germany).
3.1 Detection of subclinical mastitis
To determine the stage of subclinical mastitis directly on the farm, a mastitis detector (Draminski 4×4 Q Mast apparatus, Poland) was used. Based on the results of measuring the electrical resistance of milk, it was determined that out of 643 milk samples, 278 samples were at the stage of subclinical mastitis (Table 2). The percentage of cases of subclinical mastitis was 23 %.
Based on the data in Table 2, we can conclude that the predominant number of samples positive for subclinical mastitis was detected in the winter period (51.1 %). As a result, S. aureus isolates were more often detected in winter, so 60.9 % of isolates were detected in winter, while 39.1 % of isolates were detected in spring.
3.2 Isolation of S. aureus strains
The isolated bacteria from bovine mastitis were identified by conventional methods. According to our results, 64 isolates of S. aureus were identified in tests for catalase, plasma coagulase, and deoxyribonuclease (DNase) and also had haemolytic activity (Fig. 1). All of the 64 isolates of S. aureus were found to be gram-positive cocci.

Figure 1Tests for identification of S. aureus from subclinical bovine mastitis milk sample. (a) Tube coagulase test, (b) deoxyribonuclease test, and (c) haemolytic activity.
All 64 S. aureus isolates were positive in the biochemical identification test (STAPHYtest 24). Among 64 S. aureus strains, 16 were isolated from Simmental cows and 48 from black-and-white cows.
3.3 Antimicrobial susceptibility testing
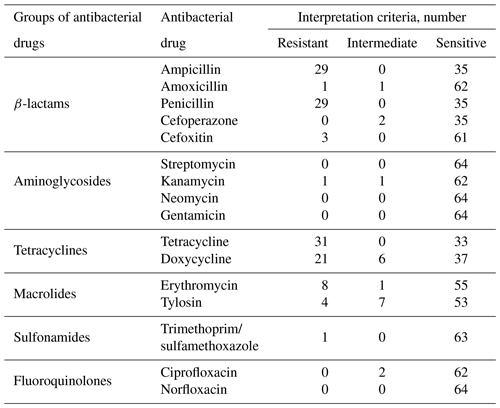
According to the results of antibiotic susceptibility testing, the greatest resistance was observed to tetracycline (48.4 %), ampicillin (45.3 %), penicillin (45.3 %), and doxycycline (32.8 %) (Table 3).
Among the studied microorganisms, cases of multidrug resistance (Magiorakos et al., 2012) were registered: three strains of microorganisms were resistant to three groups of antimicrobial drugs and two strains to four groups. All studied strains were susceptible to cefoperazone, streptomycin, neomycin, gentamicin, ciprofloxacin, and norfloxacin.
3.4 Testing the ability to form biofilms
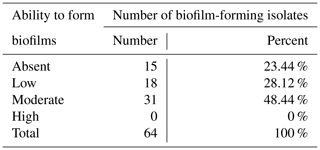
Among studied S. aureus isolates, 76.6 % () had the ability to form biofilms (Table 4).
Among biofilm-forming strains, 69.4 % () were resistant to at least one antimicrobial agent. We observed that 26 of 49 biofilm-forming strains were resistant to tetracycline, ampicillin (), penicillin (), and doxycycline ().
In addition, 43.75 % of the studied samples were simultaneously resistant to one or more antibiotics, had the ability to form a biofilm, and were a carrier of the resistance gene. In contrast, 12.5 % of isolates were resistant and formed biofilms, 4.7 % carried the resistance gene and formed biofilms, and 10.9 % did not form biofilms but were resistant and carried antibiotic resistance genes.
3.5 Detection of resistance genes
The genes for resistance to β-lactams (blaZ, mecA, msrA), macrolides (ermC), aminoglycosides (aph (3)), and tetracyclines (tetK, tetM) were detected by PCR (Table 5).
The results have shown that 46.9 % (30/64) of S. aureus isolates expressed the blaZ gene, of which 83.3 % () were resistant to penicillin, and 16.7 % () were susceptible. The presence of the mecA gene was found in one DNA isolate resistant to β-lactams (ampicillin). The ermC gene encoding resistance to macrolides was expressed in 23.4 % isolates (). It should be noted that only two DNA samples () had phenotypic resistance to the tested antibiotics of the macrolide group (erythromycin, tylosin). Both DNA isolates carrying genes for resistance to aminoglycosides – aph (3) – were sensitive to antibiotics of this group. Among the tested DNA isolates of S. aureus, 19 expressed the tetK gene, 9 expressed the tetM gene, and all were phenotypically resistant to drugs of the tetracycline group.
S. aureus is an important etiological factor in the development and spread of subclinical mastitis. Subclinical mastitis causes serious economic losses associated with reduced milk production and poor animal health (Patel et al., 2021). The high prevalence of a specific pathogen jeopardizes efforts to contain the spread of mastitis in dairy cattle (Cheng et al., 2019). In the present study, S. aureus was isolated from 23.02 % () of raw milk samples taken from dairy cows at the stage of subclinical mastitis. The above results are 2.8 % lower than the reported prevalence in China (Liu et al., 2017) and 10.6 % higher than similar studies in Iran (Jamali et al., 2015). However, the real result was lower than that obtained in some countries such as China (32 %; Liu et al., 2017) and Brazil (27.4 %; Rall et al., 2014). It should also be noted that the detection rate of S. aureus in a milk sample at the stage of subclinical mastitis was higher in similar studies in some countries such as Italy (9.1 %; Riva et al., 2015) and USA (2.9 %; Pol et al., 2007). In general, in recent years, the percentage of S. aureus isolated from dairy cows' milk samples at the stage of subclinical mastitis varied from 12 to 46 % (Jamali et al., 2015; Wang et al., 2018).
According to our research, it was found that S. aureus isolates were most often isolated in the winter months (60.9 %). This may be due to seasonal differences, as according to Koivula (2007) contagious mastitis pathogens such as S. aureus are more common in cow barns. The study of the seasonal prevalence of mastitis pathogens was not our main goal, but many researchers have studied this problem. Karzis et al. (2019) noted that in the warm months the number of cows with mastitis increases, which is caused by high humidity and poor hygiene of pens and bedding. However, it has also been reported (Matallah et al., 2019) that crowded indoor conditions during winter also affect the occurrence of the mastitis pathogen. In the present study, most of the resistant strains were isolated during the winter months as 76.9 % of winter isolates were resistant to one or more classes of antibacterial agents. A high level of resistance in the winter months was reported for β-lactam drugs; in particular, 73 % of resistant strains were resistant to ampicillin, and 76 % were resistant to penicillin. In the spring months, the number of resistant strains was 52 %. The majority of them were tetracycline-resistant (tetracycline 92 %, doxycycline 76.9 %). Our results are consistent with data obtained by other researchers from the USA and Norway (Makovec et al., 2003; Belmamoun et al., 2016; Karzis et al., 2019; Matallah et al., 2019; Saeed et al., 2022).
In our study, 48.4 % of the strains showed resistance to tetracycline, 45.3 % to ampicillin and penicillin, and 32.8 % to doxycycline. S. aureus is most commonly resistant to ampicillin and penicillin, which is consistent with previous studies in Malaysian states (Saeed et al., 2022). S. aureus bacteria were also isolated at 37 ∘C in the article by Saeed et al. (2022). The predominant number of tetracycline-resistant strains may be associated with the widespread use of this antibiotic for the treatment and prevention of many infections (Jamali et al., 2015).
The high level of resistance to ampicillin and penicillin among S. aureus isolates in most cases is associated with the use of these drugs for the prevention and treatment of mastitis (Saini et al., 2012; Rabello et al., 2005). According to the register of veterinary drugs of the Republic of Kazakhstan, β-lactam drugs are most often used to treat animals (State Register of Veterinary Drugs, 2021). The staphylococcal isolates studied in this work had phenotypic resistance to the tested antibacterial drugs used for a certain period of time. Phenotypic resistance data were confirmed by molecular genetic methods of research. Thus, the BlaZ gene encoding the enzyme β-lactamase was found in 46.9 % of resistant isolates. The BlaZ gene is widely distributed among S. aureus isolates isolated from milk at the stage of subclinical mastitis (Haveri et al., 2008; Souza et al., 2019).
Multiple-drug resistance manifested itself in resistance to two, three, and four groups of antibacterial drugs at once. We observed that all studied strains were susceptible to cefoperazone, streptomycin, neomycin, gentamicin, ciprofloxacin, and norfloxacin.
Determination of biofilm formation by staining on microtiter plates is the gold standard for phenotypic biofilm analysis. Among the known methods for determining the biofilm-forming capacity of S. aureus, researchers reported the possibility of obtaining a higher level of positive results using Congo red agar plates (Dhanawade et al., 2010; Aslantaş and Demir, 2016). However, many researchers also do not recommend this method for the detection of biofilms due to the strong dependence between growth conditions and biofilm formation (Mathur et al., 2006). Today, the method of genotypic identification of genes (icaA, icaD, fnbB, bap, etc.) responsible for the synthesis of biofilms is considered promising (Aslantaş and Demir, 2016). In our study, 76.6 % of S. aureus isolates had the ability to form biofilms. Our results were consistent with a number of studies conducted in Iran (Khoramian et al., 2015), Poland (Szweda et al., 2012), and the USA (Fox et al., 2005). Most of the isolates (48.4 %) were classified as “moderate producers”. Although none of the studied isolates were classified as “strong producers”, a potential risk to public health should be noted. In addition, according to Wang et al. (2018), biofilms increase the opportunity for the transmission of antibiotic resistance genes. In our study, 43.75 % of the studied samples were resistant to one or more antibiotics and simultaneously had the ability to form biofilm and expressed the resistance gene. The above mechanisms increase the opportunity for the spread of MDR (multiple-drug resistance) S. aureus.
The present study has been carried out on S. aureus isolated from dairy cows of such breeds as Simmental and black-and-white. According to the results, 12.5 % () were isolated from Simmental cows and 9.3 % () from black-and-white cows. Therefore, we did not find significant differences in susceptibility to S. aureus in the studied breeds of cows. In the territory of northern Kazakhstan, these breeds of cows are most often used to obtain milk, as they are capable of rapid acclimatization without loss of productive qualities. Mastitis, being a production disease, depends on many factors, such as feeding, housing, immunity of the animal, endocrine, and metabolic and physiological changes before and after calving.
Examination of the antimicrobial resistance genes' expression has shown that 60.9 % () of S. aureus isolates carried between one and five resistance genes. Thus, in 46.9 % of the studied DNA isolates, the blaZ gene was found, which encodes the β-lactamase enzyme activating the antibiotic due to the hydrolysis of the peptide bond in the β-lactam ring (Jensen et al., 2009). However, not all phenotypically penicillin-resistant isolates expressed the blaZ gene. However, the above isolates had the ability to form biofilm, so it can be assumed that this resistance mechanism worked in this case.
In the present study, one S. aureus isolate was found carrying the mecA gene, which determines methicillin resistance and encodes a low-affinity penicillin-binding protein (PBP). In our previous studies conducted in 2018–2019, not a single case of methicillin-resistant staphylococcus aureus (MRSA) was registered (Chuzhebaeva et al., 2020). However, researchers from several countries reported varied levels of MRSA ranging from 0.7 % to 90 % (Keyvan et al., 2020).
Genotypic resistance to macrolides was observed by the level of ermC gene expression in the investigated isolates (23.4 %). The data were consistent with the results presented by Srednik et al. (2018). Since 13 out of 15 samples carrying the ermC gene were sensitive to drugs of the macrolide group (erythromycin, tylosin), it can be assumed that the expression of methylases encoded by the ermC gene was not significant (Gao et al., 2012). However, other authors reported more frequent resistance encoded by the ermC and msrA genes. According to Argudin et al. (2011), in Poland, the ermA, ermB, and ermC genes were detected in 70 % of cases (Argudin et al., 2011). According to Elad et al. (2012), the ermB and ermC genes were found in 100 % and 84.6 %, respectively.
The aph (3) gene responsible for resistance to aminoglycosides due to the release of aminoglycoside phosphotransferase enzymes was found in two staphylococcal DNA isolates. Also, these isolates were multi-resistant. Except for the aph (3) gene, one isolate carried genes for resistance to macrolides (ermC) and β-lactams (blaZ), and the second isolate also carried genes for tetracyclines (tetK, tetM; Omwenga et al., 2020). It can be assumed that resistance plasmids on which resistance genes are found can carry one or more additional resistance genes (Wendlandt et al., 2021).
The data obtained from the study of antibiotic resistance genes of the tetracyclines group coincided with the phenotypic studies. The tetK gene was found in 19 isolates, whereas the tetM gene was expressed in 9 isolates. In the present study, the tetK gene was found in 29.7 % of the samples and tetM in 14 %. In contrast, according to Yang et al. (2016) the percentage of expression for tetK and tetM was 22.73 % and 2.27 %, respectively.
Our earlier studies conducted on 68 isolates confirmed the presence of tetM and tetK genes in only two and three isolates, respectively (Chuzhebaeva et al., 2020). The results of the current study have shown that the level of isolation of resistance genes increased significantly. The relatively high prevalence of the tetK and tetM genes in the studied staphylococci indicated the presence of the following resistance mechanisms: tetracycline efflux pump and a ribosomal protection protein (Rahimi et al., 2015).
The development and implementation of new methods for the treatment of subclinical mastitis require separate studies. However, because of the increasing level of antibiotic resistance, special attention should be paid to the prevention of subclinical mastitis. Particular attention should be paid to the disinfection of the teats before and after milking, conditions of the animals, minimization of stress, and monitoring of the correct operation of the milking machines. Continuous monitoring of somatic cell levels is also recommended. Antibacterial susceptibility testing is recommended when an infectious agent is detected. Antimicrobial treatment of mastitis in animals should comply with the OIE Standards for Responsible and Prudent Use as set out in chap. 6.9 of the Sanitary Code for Terrestrial Animals. Veterinarians should also responsibly use antimicrobials important for veterinary medicine, such as third- and fourth-generation cephalosporins and novobiocin (World Organisation for Animal Health, 2021).
According to the data obtained, we found the presence of antimicrobially resistant isolates of S. aureus, which in turn is of concern since subclinical mastitis not only harms the animal body but also negatively affects the human body. Further detailed studies are needed to determine how dairy cattle become infected with S. aureus isolates and how the transmission of these bacteria occurs between individuals.
Our study provided that S. aureus isolates obtained from cows' milk samples at the stage of subclinical mastitis were resistant to many antibiotics of the tetracyclines and β-lactam groups which are commonly used to treat mastitis. In addition, the ability to form biofilms increased the opportunity for the spread of multidrug-resistant strains. Moreover, we confirmed that S. aureus isolates express several resistance genes which encode resistance to β-lactams and tetracyclines. In conclusion, S. aureus in raw milk is a risk factor for the development of foodborne infections and the spread of antibiotic resistance between species.
The data are available from the corresponding author upon request.
RR, NM, and ZB conceptualized the hypotheses and design of the study. AM and PS analysed the data. PS managed the figures. RR wrote the paper. JM, BM, and BO reviewed and revised the manuscript. All authors have read and agreed to the published version of the paper.
The contact author has declared that none of the authors has any competing interests.
All activities involving animals are carried out in compliance with biosafety and animal welfare standards. A positive conclusion of the local ethical commission of the Scientific Research Institute of Applied Biotechnology of the A. Baitursynov Kostanay Regional University was received for conducting experiments with animals (protocol no. 1 dated 19 May 2020).
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research has been funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (grant no. AP08956574).
This research has been funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (grant no. AP08956574).
This paper was edited by Henry Reyer and reviewed by four anonymous referees.
Ali Gharieb, R. M., Saad, M. F., Mohamed, A. S., and Tartory, Y. H.: Characterization of two novel lytic bacteriophages for reducing biofilms of zoonotic multidrug-resistant Staphylococcus aureus and controlling their growth in milk, LWT, 124, 109–145, https://doi.org/10.1016/j.lwt.2020.109145, 2020.
Argudin, M. A., Tenhagen, B. A., Fetsch, A., Sachsenroder, J., Kasbohrer, A., Schroeter, A., Hammerl, J. A., Hertwig, S., Helmuth, R., Braunig, J., Mendoza, M. C., Appel, B., Rodicio, M. R., and Guerra, B.: Virulence and Resistance Determinants of German Staphylococcus aureus ST398 Isolates from Nonhuman Sources, Appl. Environ. Microb., 77, 3052–3060, https://doi.org/10.1128/aem.02260-10, 2011.
Aslantaş, Ö. and Demir, C.: Investigation of the antibiotic resistance and biofilm-forming ability of Staphylococcus aureus from subclinical bovine mastitis cases, J. Dairy Sci., 99, 8607–8613, https://doi.org/10.3168/jds.2016-11310, 2016.
Barkema, H. W., Schukken, Y. H., and Zadoks, R. N.: Invited Review: The Role of Cow, Pathogen, and Treatment Regimen in the Therapeutic Success of Bovine Staphylococcus aureus Mastitis, J. Dairy Sci., 89, 1877–1895, https://doi.org/10.3168/jds.S0022-0302(06)72256-1, 2006.
Belmamoun, A. R., Reguig, K. B., Bouazza, S., Dif, M. M.: Subclinical mastitis on the raw milk as a risk factor for the transmission of Staphylococcus aureus and coagulase-negative staphylococci, multidrug resistance in Sidi Bel Abbes, Algeria, Adv. Environ. Biol., 10, 1–11, https://www.thefreelibrary.com/Subclinical+mastitis+on+the+raw+milk+as+a+risk+factor+for+the...-a0466412058 (last access: 4 December 2022), 2016.
Cheng, J., Qu W., Barkema, H. W., Nobrega, D. B., Gao, J., Liu G., De Buck, J., Kastelic, J. P., Sun, H., and Han, B.: Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds, J. Dairy Sci., 102, 2416–2426, https://doi.org/10.3168/jds.2018-15135, 2019.
Chuzhebaeva, G., Baimenov, B., Aliyeva, G., Siugzdiniene, R., Mendybayeva, A., and Rychshanova, R.: Study of the antibacterial sensitivity of staphylococcus aureus isolated from dairy cattle, Ecol. Envir. Con., 26, 1688–1692, 2020.
CLSI: Performance Standards for Antimicrobial Susceptibility Testing, 29th edn., CLSI supplement M100, Clinical and Laboratory Standards Institute, Wayne, PA, 2019.
Dhanawade, N. B., Kalorey, D. R., Srinivasan, R., Barbuddhe, S. B., and Kurkure, N. V.: Detection of intercellular adhesion genes and biofilm production in Staphylococcus aureus isolated from bovine subclinical mastitis, Vet. Res. Commun., 34, 81–89, 2010.
Elad, D., Blum, S., Fleker, M., Zukin, N., Weissblit, L., and Shlomovitz, S.: Analysis of Long Term (20 Years) In Vitro Susceptibility to Antibacterial Drugs of the Most Prevalent Animal Bacterial Pathogens Isolated in Israel, Part 2: Multi-Drug Resistance, Isr. J. Vet. Med., 67, 133–138, 2012.
EUCAST – European Committee on Antimicrobial Susceptibility Testing: Breakpoint tables for interpretation of MICs and zone diameters, Version 11.0, valid from 2021/01-01, https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (last access: 21 November 2022), 2021.
Fox, L. K., Zadoks, R. N., and Gaskins, C. T.: Biofilm production by Staphylococcus aureus associated with intramammary infection, Vet. Microbiol., 107, 295–299, https://doi.org/10.1016/j.vetmic.2005.02.005, 2005.
Gaglio, R., Couto, N., Marques, C., Lopes, M., Moschetti, G., Pomba, C., and Settanni, L.: Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses, Intern. J. Food. Microb., 236, 107–114, https://doi.org/10.1016/j.ijfoodmicro.2016.07.020, 2016.
Gao, J., Ferreri, M., Yu, F., Liu, X., Chen, L., Su, J., and Han, B.: Molecular types and antibiotic resistance of Staphylococcus aureus isolates from bovine mastitis in a single herd in China, Vet. J., 192, 550–552, https://doi.org/10.1016/j.tvjl.2011.08.030, 2012.
Haveri, M., Hovinen, M., Roslof, A., and Pyorala, S.: Molecular Types and Genetic Profiles of Staphylococcus aureus Strains Isolated from Bovine Intramammary Infections and Extramammary Sites, J. Clin. Microbiol., 46, 3728–373, https://doi.org/10.1128/jcm.00769-08, 2008.
Hennekinne, J. A., De Buyser, M. L., and Dragacci, S.: Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation, FEMS Microb. Rev., 36, 815–836, https://doi.org/10.1111/j.1574-6976.2011.00311.x, 2012.
Jamali, H., Paydar, M., Radmehr, B., Ismail, S., and Dadrasnia, A.: Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products, Food Contr., 54, 383–388, https://doi.org/10.1016/j.foodcont.2015.02.013, 2015.
Jensen, A. G., Wachmann, C. H., Espersen, F., Scheibel, J., Skinhoj, P., and Frimodt, M.: Treatment and Outcome of Staphylococcus aureus Bacteremia. A prospective study of 278 Cases, Arch. Intern. Med., 162, 25–32, https://doi.org/10.1001/archinte.162.1.25, 2002.
Jensen S. O. and Lyon B. R.: Genetics of antimicrobial resistance in Staphylococcus aureus, Future Microbiol., 4, 565–82, https://doi.org/10.2217/fmb.09.30, 2009.
Karzis, J., Petzer, I. M., Donkin, E. F., Naidoo, V., and Etter, E. M. C.: Climatic and regional antibiotic resistance patterns of Staphylococcus aureus in South African dairy herds, Onder. J. Vet Res, 86, a1674, https://doi.org/10.4102/ojvr.v86i1.1674, 2019.
Keyvan, E., Yurdakul, O., Demirtas, A., Yalcin, H., and Bilgen, N.: Identification of Methicillin-Resistant Staphylococcus aureus in Bulk Tank Milk, Food Sci. Techn., 40, 150–156, https://doi.org/10.1590/fst.35818, 2020.
Khoramian, B., Jabalameli, F., Niasari-Naslaji, A., Taherikalani, M., and Emaneini, M.: Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections, Mic. Pathog., 88, 73–77, https://doi.org/10.1016/j.micpath.2015.08.007, 2015.
Klimiene, I., Virgailis, M., Pavilonis, A., Siugzdiniene, R., Mockeliunas, R., and Ruzauskas, M.: Phenotypical and genotypical antimicrobial resistance of coagulase-negative staphylococci isolated from cow mastitis, Pol. J. Vet. Sci., 19, 639–646, https://doi.org/10.1515/pjvs-2016-0080, 2016.
Koivula, M., Pitkälä, A., Pyörälä, S., and Mäntysaari, E. A.: Distribution of bacteria and seasonal and regional effects in a new database for mastitis pathogens in Finland, Acta Agr. Scand. A-An., 57, 89–96, https://doi.org/10.1080/09064700701488941, 2007.
Liu, H., Li, S., Meng, L., Dong, L., Zhao, S.H., Lan, X., Wang, J., and Zheng, N.: Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China, J. Dairy Sci., 100, 8796–8803, https://doi.org/10.3168/jds.2017-13370, 2017.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., and Monnet, D. L.: Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance, Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 18, 268–81, https://doi.org/10.1111/j.1469-0691.2011.03570.x, 2012.
Makovec, J. A. and Ruegg, P. L.: Results of Milk Samples Submitted for Microbiological Examination in Wisconsin from 1994 to 2001, J. Dairy Sci., 86, 3466–3472, https://doi.org/10.3168/jds.s0022-0302(03)73951-4, 2003.
Matallah, A. M., Leila Bouayad, L., Boudjellaba, S., Mebkhout, F., Hamdi, T. M., and Ramdani-Bouguessa, N.: Staphylococcus aureus isolated from selected dairies of Algeria: Prevalence and susceptibility to antibiotics, Vet World, 12, 205–2010, https://doi.org/10.14202/vetworld.2019.205-210, 2019.
Mathur T., Singhal S., Khan S., Upadhyay D.J., Fatma T., and Rattan A. Detection of biofilm formation among the clinical isolates of staphylococci: an evaluation of three different screening methods, Indian J. Med. Microbi., 24, 25–29, https://doi.org/10.1016/S0255-0857(21)02466-X, 2006.
MI 4.2.1890-04: Determination of the sensitivity of microorganisms to antibacterial drugs. Methodical instructions, M.: FCHE Rospotrebnadzor. Introduced from 04.03, https://antimicrob.net/wp-content/uploads/4.pdf (last access: 21 November 2022), 2004.
Oliver, S. P., Gonzalez, R. N., Hogan, J. S., Jayarao, B. M., and Owens, W. E.: Microbiological procedures for the diagnosis of bovine udder infection and determination of milk quality, National Mastitis Council, Verona, 47, https://www.worldcat.org/title/microbiological-procedures-for-the-diagnosis-of-bovine-udder-infection-and-determination-of-milk-quality/oclc/61200882 (last access: 4 December 2022), 2004.
Okulich, V. K.: Microbial biofilms in clinical microbiology and antibacterial therapy, edited by: Okulich, V. K., Kabanova, A. A., and Plotnikov, F. V., Vitebsk: VSMU, pp. 136–137, https://www.elib.vsmu.by/bitstream/123/12846/1/Okulich-VK_Mikrobnye_bioplenki_v_klinicheskoj_mikrobiologii_i_antibakterial%27noj_terapii_2017.pdf (last access: 21 November 2022), 2017 (in Russian).
Omwenga, I., Aboge, G. O., Mitema, E. S., Obiero, G., Ngaywa, C., Gaywa, C., Ngwili, N., Wamwere, G., Wainaina, M., and Bett, B.: Antimicrobial usage and detection of multidrug-resistant Staphylococcus aureus, including methicillin-resistant strains in raw milk of livestock from Northern Kenya, Microb. Drug Resist., 27, 843–854, https://doi.org/10.1089/mdr.2020.0252, 2020.
Patel, K., Godden, S. M., Royster, E. E., Crooker, B. A., Johnson, T. J., Smith, E. A., and Sreevatsan, S.: Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of U.S. dairy herds, BMC Genomics 22, 367, https://doi.org/10.1186/s12864-021-07603-4, 2021.
Pol, M. and Ruegg, P. L.: Relationship Between Antimicrobial Drug Usage and Antimicrobial Susceptibility of Gram-Positive Mastitis Pathogens, J. Dairy Sci., 90, 262–273, https://doi.org/10.3168/jds.S0022-0302(07)72627-9, 2007.
Rabello, R. F., Souza, C. R. V. M., Duarte, R. S., Lopes, R. M. M., Teixeira, L. M., and Castro, A. C. D.: Characterization of Staphylococcus aureus Isolates Recovered from Bovine Mastitis in Rio de Janeiro, Brazil, J. Dairy Sci., 88, 3211–3219, https://doi.org/10.3168/jds.S0022-0302(05)73004-6, 2015.
Rahimi, H., Dastmalchi, S. H., and Ahmadi, M.: Nasal Carriage of Staphylococcus aureus: Frequency and Antibiotic Resistance in Healthy Ruminants, Jundishapur J. Microbiol., 8, e22413, https://doi.org/10.5812/jjm.22413, 2015.
Rall, V. L. M., Miranda, E. S., Castilho, I. G., Camargo, C. H., Langoni, H., Guimarães, F. F., Araújo Júnior, J. P., and Fernandes Júnior, A.: Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis, J. Dairy Sci., 97, 829–837, https://doi.org/10.3168/jds.2013-7226, 2014.
Riva, A., Borghi, E., Cirasola, D., Colmegna, S., Borgo, F., Amato, E., Pontello, M. M., and Morace G.: Methicillin-Resistant Staphylococcus aureus in Raw Milk: Prevalence, SCCmec Typing, Enterotoxin Characterization, and Antimicrobial Resistance Patterns, J. Food Prot., 78, 1142–1146, https://doi.org/10.4315/0362-028X.JFP-14-531, 2015.
Ruzauskas, M., Siugzdiniene, R., Klimiene, I., Virgailis, M., Mockeliunas, R., Vaskeviciute, L., and Zienius, D.: Prevalence of methicillin-resistant Staphylococcus haemolyticus in companion animals: a cross-sectional study, Ann. Clin. Microbiol. Antimicrob., 13, 56, https://doi.org/10.1186/s12941-014-0056-y, 2014.
Ruzauskas, M., Couto, N., Pavilonis, A., Klimiene, I., Siugzdiniene, R., Virgailis, M., Vaskeviciute, L., Anskiene, L., and Pomba, C.: Characterization of Staphylococcus pseudintermedius isolated from diseased dogs in Lithuania, Pol. J. Vet. Sci., 19, 7–14, https://doi.org/10.1515/pjvs-2016-0002, 2016.
Saeed, S. I., Mat Yazid, K. A., Hashimy, H. A., Dzulkifli, S. K., Nordin, F., Nik Him, N. A., Omar, M. F. F. B., Aklilu, E., Mohamad, M., Zalati, C. W. S., and Kamaruzzaman, N. F.: Prevalence, Antimicrobial Resistance, and Characterization of Staphylococcus aureus Isolated from Subclinical Bovine Mastitis in East Coast Malaysia, Animals, 12, 1680, https://doi.org/10.3390/ani12131680, 2022.
Saini, V., McClure, J. T., Scholl, D. T., DeVries, T. J., and Barkema, H. W.: Herd-level association between antimicrobial use and antimicrobial resistance in bovine mastitis Staphylococcus aureus isolates on Canadian dairy farms, J. Dairy Sci., 95, 1921–1929, https://doi.org/10.3168/jds.2011-5065, 2012.
Shathele, M. S.: Weather effect on bacterial mastitis in dairy cows, Inter. J. Dairy. Sci., 4, 57–66, https://doi.org/10.3923/ijds.2009.57.66, 2009.
Souza, G., de Almeida, A. C., Xavier, M., da Silva, L., Sousa, C. N., Sanglard, D. A., and Xavier, A.: Characterization and molecular epidemiology of Staphylococcus aureus strains resistant to beta-lactams isolated from the milk of cows diagnosed with subclinical mastitis, Vet. World, 12, 1931–1939, https://doi.org/10.14202/vetworld.2019.1931-1939, 2019.
Srednik, M. E., Usongo, V., Lepine, S., Janvier, X., Archambault, M., and Gentilini, E. R.: Characterization of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina, J. Dairy Res. 85, 57–63, https://doi.org/10.1017/S0022029917000851, 2018.
State Register of Veterinary Drugs and Feed Additives: Ministry of Agriculture of the Republic of Kazakhstan. Committee for Veterinary Control and Supervision, Nur-Sultan, https://online.zakon.kz/Document/?doc_id=31651085&pos=5;-106#pos=5;-106 (last access: 21 November 2022), 2021.
State Standard 30347-2016: Milk and dairy products. Methods for the determination of Staphylococcus aureus, Moscow Standard inform., p. 14, https://internet-law.ru/gosts/gost/63606/ (last access: 21 November 2022), 2016.
Szweda, P., Schielmann, M., Milewski, S., Frankowska, A., and Jakubczak, A.: Biofilm Production and Presence of ica and bap Genes in Staphylococcus aureus strains isolated from cows with mastitis in the Eastern Poland, Pol. J. Microb, 61, 65–69, https://doi.org/10.33073/pjm-2012-009, 2012.
World Organisation for Animal Health: Terrestrial Animal Health Code, 29th edn., Vol. 1, Chap. 6.9, General provisions, https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/?id=169&L=1&htmfile=chapitre_antibio_monitoring.htm (last access: 21 November 2022), 2021.
Thiran, E., Di Ciccio, P. A., Graber, H. U., Zanardi, E., Ianieri, A., and Hummerjohann, J.: Biofilm formation of Staphylococcus aureus dairy isolates representing different genotypes, J. Dairy Sci., 101, 1000–1012, https://doi.org/10.3168/jds.2017-13696, 2018.
Timakov, A. V., Timakova, T. K., and Shmarov, A. T.: Staphylococcal mastitis and food safety of milk and dairy products, Bulletin of the agro-industrial complex of the Upper Volga Region, 4, 56–59, https://yaragrovuz.ru/images/Vestnik_APK/15-4/56-59_4-2015.pdf (last access: 21 November 2022), 2015 (in Russian).
Wang, W., Lin, X., Jiang, T., Peng, Z., Xu, J., Yi, L. F., Fanning, S., and Baloch, Z.: Prevalence and Characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China, Front. Microb., 9, 1123, https://doi.org/10.3389/fmicb.2018.01123, 2018.
Wendlandt S., Feßler A. T., Monecke S., Ehricht R., Schwarz S., and Kadlec K.: The diversity of antimicrobial resistance genes among staphylococci of animal origin, Int. J. Med. Microbiol., 303, 338–349, https://doi.org/10.1016/j.ijmm.2013.02.006, 2021.
Yang, F., Wang, Q., Wang, X., Wang, L., Xiao, M., Li, X., Luo, J., Zhang, S., and Li, H.: Prevalence of blaZ gene and other virulence genes in penicillin-resistant Staphylococcus aureus isolated from bovine mastitis cases in Gansu, China, Turk. J. Vet. Anim Sci., 39, 634–636, https://doi.org/10.3906/vet-1504-81, 2015.
Yang, F., Wang, Q., Wang, X., Wang, L., Li, X., Luo, J., Zhang, S. H., and Li, H.: Genetic characterization of antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis cases in Northwest China, J. Integr. Agric., 15, 2842–2847, https://doi.org/10.1016/S2095-3119(16)61368-0, 2016.