the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Variation in bovine leptin gene affects milk fatty acid composition in New Zealand Holstein Friesian × Jersey dairy cows
Ishaku Lemu Haruna
Huitong Zhou
Jon G. H. Hickford
Leptin is a protein hormone secreted from white adipose tissue. It regulates food/feed intake, body weight, immune function and reproduction. In our investigation, the polymerase chain reaction (PCR) amplification coupled with single-strand conformational polymorphism (SSCP) analysis was used to reveal variation in bovine leptin gene (LEP) in New Zealand (NZ) Holstein Friesian × Jersey (HF × J) dairy cows. Subsequent sequence analysis of a 430 bp amplicon spanning the entirety of exon 3 and part of the intron 2 region revealed three variant sequences (A3, B3 and C3) containing a total of five nucleotide substitutions, all of which have been reported previously. Using general linear mixed-effect model analyses, the presence of variant A3 (the most common variant) was associated with a decreased level of C15:1, C18:1 trans-11, C18:1 all trans, C18:2 trans-9, cis-12, C22:0 and C24:0 levels but increased levels of C12:1 and C13:0 iso (p<0.05). Variant B3 was associated with reduced levels of C6:0, C8:0, C11:0, C13:0 and C20:0 but increased C17:0 iso and C24:0 levels (p<0.05). Variant C3 was associated with decreased C17:0 iso levels but increased C20:0 (p<0.05) levels. In a genotype model, the A3B3 genotype was associated with increased levels of C22:0 and C24:0 but decreased C8:0, C10:0, C11:0, C13:0, C15:0 and grouped medium-chain fatty acid (MCFA) levels (p<0.05). Genotype A3C3 was found to be associated with decreased levels of C10:0, C11:0, C13:0 and grouped MCFA (p<0.05). This is the first report of findings of this kind in NZ HF × J cows, and they suggest that variation in exon 3 of bovine leptin gene could be explored as a means of decreasing the concentration of saturated fatty acids in milk.
- Article
(329 KB) - Full-text XML
- BibTeX
- EndNote
There has been a growing interest in genomic selection programmes aimed at modifying the composition of milk fatty acids (FAs) using candidate gene approaches. In this respect, several genes have been implicated in affecting milk FA composition, including the leptin gene (LEP).
Bovine LEP, previously known as OB, OBS and LEPD, has been mapped to chromosome 4 (Pomp et al., 1997) and it encodes the protein leptin. This protein is secreted from white adipose tissue and has been found to regulate feed intake, energy partitioning and metabolism (Liefers et al., 2002; Lagonigro et al., 2003), as well as lactogenesis (Feuermann et al., 2004).
The hypothalamus is identified as the main site of leptin's activity in regulating food intake and energy expenditure. Leptin signals are converted into neural responses, and this results in changes in feed intake (Tang-Christensen et al., 1999). A neurotransmitter identified as neuropeptide Y (NPY) is associated with the regulation of food intake, and leptin exerts its effect by either stimulating or inhibiting the release of NPY. Among other things, this eventually results in a decrease in feed intake and an increase in energy expenditure (Houseknecht et al., 1998). There are also suggestions that leptin could also regulate fat mobilization (Halaas et al., 1995).
Previous reports have highlighted the effects of leptin gene variation on some livestock traits of economic value, such as the yield and quality of meat and milk obtained from farmed animals. For example, in sheep an effect of leptin gene variation on weaning weight was observed (Hajihosseinlo et al., 2012), while in cattle, leptin or leptin receptor gene polymorphisms have been associated with carcass FA composition (Kawaguchi et al., 2017), milk fat levels (Giblin et al., 2010; De Matteis et al., 2012) and milk FA composition (Pegolo et al., 2016).
Although the effects of bovine leptin variation on milk fat composition have been described in studies of other cattle breeds, so far there is no report of the effects of leptin gene variation on the composition of milk FA content or profile in New Zealand (NZ) Holstein Friesian × Jersey (HF × J) dairy cows that are permanently grazed outdoors on pasture. The aim of this study was therefore to investigate whether variation in the gene affected milk fat traits in these cows.
2.1 The NZ dairy cattle investigated
This study was approved by the Lincoln University Animal Ethics Committee (AEC) under the provisions of the NZ Government's Animal Welfare Act 1999. A total of 300 NZ HF × J dairy cows (alternatively known as KiwiCross™ cows) of variable and unknown breed proportion and of 3 to 9 years of age were used in this investigation. These cows were from two herds, and all of them were grazed outdoors on pasture (a mixture of perennial ryegrass and white clover) on the Lincoln University Dairy Farm (LUDF; Canterbury, NZ). All the cows calved over the months of August–September and they were milked twice a day, in the morning and then in the afternoon.
2.2 Collection of milk samples for fatty acid analysis
The collection of milk samples from cows for FA analysis was carried out when they were 148 ± 19 d in milk (DIM) and in a single afternoon milking in mid-January. These samples were frozen at a temperature of −20∘C, and then freeze-dried, before being individually ground to a fine powder for component analysis.
2.3 Gas chromatography of milk fatty acids
Prior to being analysed by gas chromatography (GC) as FA methyl esters (FAMEs), the milk FAs were methylated and then extracted in n-heptane. The methylation reactions were performed in 10 mL Kimax tubes. Individual freeze-dried and powdered milk samples (0.17 g) were dissolved in 900 µL of n-heptane (100 %, AR grade), before 100 µL of internal standard (5 mg/mL of C21:0 methyl ester in n-heptane) and 4.0 mL of 0.5 M NaOH (in 100 % anhydrous methanol) were added.
The tubes were vortexed prior to incubation in a block heater (Ratek Instruments, Australia) at 50∘C for 15 min. After cooling to room temperature, another 2.0 mL of n-heptane and 2.0 mL of deionized water were added to each of the tubes. After vortexing, the tubes were centrifuged (Megafuge 1.0R, Heraeus, Germany) for 5 min at 1500× g. The top layer of n-heptane was transferred into a second Kimax tube, and 2.0 mL of n-heptane was added to each of the original tubes. The extraction was repeated, and the n-heptane aspirates were then pooled. Anhydrous sodium sulfate (10 mg) was added to the n-heptane extracts, to remove any residual water.
The GC analysis for milk FAs was carried out using a Shimadzu GC-2010 gas chromatograph (Shimadzu Corporation, Kyoto, Japan) equipped with a flame ionization detector and an AOC-20i autosampler. The output was analysed with GC Solution Software (Shimadzu). The analysis was carried out by injecting 1 µL of the n-heptane sample extract into a 100 m GC capillary column (250 µm × 0.25 µm, CP-Select, Varian) with a 1:60 split ratio. The separation was undertaken with a helium carrier gas, and it was run for 92 min. The temperature of both the injector and detector were set at 250∘C, and the thermal profile of the column incubation consisted of 45∘C for 4 min, followed by 27 min at 175∘C (ramped at 13∘C/min), 35 min at 215∘C (ramped at 4∘C/min), and a final temperature of 250∘C for 5 min (ramped at 25∘C/min).
The individual FAMEs were identified by comparing their peak retention times to commercially obtained external standards (ME61, ME93, BR3, BR2, ME100, GLC411 and GLC463; Larodan AB, Sweden). Quantification of the individual FAMEs was based on peak area assessment and comparison with the internal and external standards. The threshold for peak area determination on the chromatogram was a 500-unit count, and peaks under this threshold were ignored. The calculated minimum level of an individual FAME that could be identified was therefore 0.01 g per 100 g of total FA. After the FAs were individually measured, they were sorted into various groups and indices. These groups were the following: short-chain FAs (SCFAs) = C4:0 + C6:0 + C8:0; medium-chain FAs (MCFA) = C10:0 + C12:0 + C14:0; long-chain FAs (LCFAs) = C15:0 + C16:0 + C17:0 + C18:0 + C19:0 + C20:0 + C22:0 + C24:0; omega-3 FAs = C18:3 cis-9, 12, 15 + C20:5 cis-5, 8, 11, 14, 17 + C22:5 cis-7, 10, 13, 16, 19; omega-6 FAs = C18:2 cis-9, 12 + C18:3 cis-6, 9, 12 + C20:3 cis-8, 11, 14 + C20:4 cis-5, 8, 11, 14; monounsaturated FAs (MUFA) = C10:1 + C12:1 + C14:1 cis-9 + C15:1 + C16:1 cis-9 + C17:1 + C18:1 trans-11 + C18:1 cis-9 + C18:1 cis-(10 to 15) + C20:1 cis-5 + C20:1 cis-9 + C20:1 cis-11 + C22:1 trans-13; polyunsaturated FAs (PUFA) = C18:2 trans-9, 12 + C18:2 cis-9, trans-13 + C18:2 cis-9, trans-12 + C18:2 trans-9, cis-12 + C18:2 cis-9, 12 + C18:3 cis-6, 9, 12 + C18:3 cis-9, 12, 15 + conjugated linoleic acid (CLA) + C20:3 cis-8, 11, 14 + C20:4 cis-5, 8, 11, 14 + C20:5 cis-5, 8, 11, 14, 17 + C22:5 cis-7, 10, 13, 16, 19; and total branched FA = C13:0 iso + C13:0 anteiso + C15:0 iso + C15:0 anteiso + C17:0 iso.
Unsaturated FA indices were also calculated as follows: C12:1 index (C12:1 divided by the sum of C12:0 and C12:1); C14:1 index (C14:1 cis-9 divided by the sum of C14:0 and C14:1 cis-9); C16:1 index (C16:1 cis-9 divided by the sum of C16:0 and C16:1 cis-9) and C18:1 index (C18:1 cis-9 divided by the sum of C18:0 and C18:1 cis-9). The method is as described by Li et al. (2019), with the un-adjusted mean levels in the 300 cows being calculated and used subsequently in the statistical analyses.
2.4 Blood sample collection
Using either the piercing of the animal's ear or the tail vein (as approved under the Code of Welfare, section 75 and 76 of the NZ Animal Welfare Act 1999), blood samples were collected from each cow onto FTA™ cards (Whatman™, Middlesex, UK). The samples were air-dried and DNA purification was carried out using a two-step procedure described by Zhou et al. (2006).
2.5 Amplification with the polymerase chain reaction (PCR)
Using the following forward and reverse primers (5′-TTGCTCTCCCCTTCCTCCTG-3′ and 5′-CTCAGGTTTCTTCCCTGGAC-3′ respectively) adapted from the work of Haruna et al. (2020), the entirety of exon 3 and part of the intron 2 region of the bovine leptin gene was amplified. This region was selected for investigation because it is highly polymorphic in comparison to the exon 2 region, and previous report has revealed associations of exon 3 with FA composition in muscle (Orrù et al., 2011). The PCR reactions were undertaken in 15 µL volumes containing the genomic DNA on a 1.2 mm diameter disc of the FTA™ card, 0.25 µM for each primer, 150 µM for each dNTP (Eppendorf, Hamburg, Germany), 3.0 mM Mg2+, 0.5 U of Taq DNA polymerase (Qiagen, Hilden, Germany), and 1× the reaction buffer supplied with the enzyme.
The amplification was carried out in Bio-Rad S1000 thermal cyclers (Bio-Rad, Hercules, CA, USA). The thermal cycling conditions included an initial denaturation at 94 ∘C for 2 min, followed by 35 cycles of 94 ∘C for 30 s, annealing for 30 s at 60 ∘C, extension at 72 ∘C for 30 s and a final extension step at 72 ∘C for 5 min.
2.6 Single-strand conformational polymorphism (SSCP) analyses
An SSCP technique was used to detect genetic variation in the amplicons obtained from the PCR reactions. The choice of SSCP was because it is inexpensive and can screen for variation in a large number of cattle breeds, thus giving a better representation of the entire breed. Also, it is a reliable, reproducible and effective analytical method for the detection of deletions, insertions or rearrangement in PCR-amplified DNA sequence. Briefly, following PCR amplification, a 0.7 µL aliquot of the PCR reactions was added to 7 µL of loading dye containing 10 mM ethylenediaminetetraacetic acid (EDTA), 0.025 % bromophenol blue, 0.025 % xylene cyanol, and 98 % formamide. The samples were then placed on a hot plate already set at 95 ∘C, for 5 min to enable DNA denaturation. This was followed by snap chilling on wet ice. Samples were then loaded onto 16 cm × 18 cm, 10 % acrylamide : bisacrylamide (37.5:1) (Bio-Rad) gels containing 4 % glycerol. Electrophoresis was carried out using Protean II xi cells (Bio-Rad) for 24 h at 390 V and 15 ∘C in 0.5× Tris/Borate/EDTA running buffer.
To detect the SSCP banding patterns, the gels were silver-stained using a method described by Byun et al. (2009).
2.7 Nucleotide sequencing
Based on the PCR-SSCP patterns observed, cattle that were homozygous with unique banding patterns were sequenced directly. For heterozygous variants, the unique band(s) was excised from the wet gel, incubated in water at 69 ∘C for 1 h, and subsequently amplified and sequenced based on the approach described by Gong et al. (2011). The sequences were then aligned, and other analyses were undertaken using DNAMAN (Version 5.2.10, Lynnon Biosoft, Vaudreuil, Canada) to enable identification of the position of the nucleotide variation.
2.8 Statistical analysis
The statistics software IBM SPSS version 22 (IBM, Armonk, NY, USA) was used to perform all statistical analyses, and an alpha level of p<0.05 was set as a threshold for acceptance of association.
The age of the cow expressed in an integer value of years (i.e. as a categorical variable in a range from 3 to 9 years of age), the number of days in milk for each cow (DIM; expressed as an integer value but entered into the model as a continuous trait) and herd (to correct for herd-specific effects) were fitted to the models as fixed explanatory factors.
Using general linear mixed-effects models (GLMMs), associations between LEP variants and variation in milk FA component levels were tested.
First, single-variant presence/absence models (each variant was coded as present (1) or absent (0) for each animal's genotype) were used to ascertain which variant(s) should be analysed in subsequent multi-variant models. The multi-variant models included any variant that had a variant-FA trait association in the single-variant presence/absence analysis with a p value of less than 0.200. This is a low threshold for the inclusion of a possibly explanatory factor in the model. The multi-variant models were also corrected for the other factors described above.
For genotypes with a frequency greater than 5 % (thus having adequate sample size per group), the effect of variation in a cow's LEP genotype on the component levels of individual and grouped FAs was tested using general linear mixed-effects models (GLMMs) and multiple pair-wise comparisons (least significant difference tests) with Bonferroni corrections.
The model was Yijkl= μ + Gi+ Aj+ Dk+ Hl+ eijkl for the genotype, where Yijkl is the observed trait value in the ijklth cow; μ is the mean trait value for a given trait; Gi is the fixed effect of ith LEP genotype; Aj is the effect of age (j=3–9 years); Dk the effect of the number of days the cow has produced milk (DIM: k=94–186 d); Hl the fixed effect of lth farm (l=1 or 2); and eijkl is the random error.
The effect of sire of cow could not be included in the GLMMs, because some semen straws (sire genetics) used in NZ dairy cattle artificial insemination-based breeding approaches contain mixed-sire semen purchased from commercial semen producers. In these cases, it is impossible to ascertain individual sire identity. However, since the straws were mixed-semen straws and because different sires are used for different inseminations, in different years, it is unlikely that sire was a strongly confounding effect. Cow age and herd might also be confounded with sire, but this cannot be confirmed.
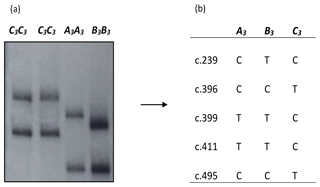
3.1 SNPs identified in the bovine leptin gene
Using the primers 5′-TTGCTCTCCCCTTCCTCCTG-3′ and 5′-CTCAGGTTTCTTCCCTGGAC-3′, a fragment of approximately 430 bp length and consisting of the entire exon 3 and part of intron 2 region of bovine leptin gene was amplified and analysed using the PCR-SSCP analyses. The PCR-SSCP analyses coupled with DNA sequencing revealed three banding patterns (A3, B3 and C3) with NCBI GenBank accession numbers MN119553, MN119554 and MN119555 respectively in the region investigated (Fig. 1). A total of five single-nucleotide substitutions – c.239C/T (p.Ala80Val), c.396C/T (p.Gly132=), c.399T/C (p.Val133=), c.411T/C (p.Ala137=) and c.495C/T (p.Pro165=) in exon 3 – were identified, all of which have been reported previously (Haruna et al., 2020).
3.2 Variant presence/absence models
The results of the general linear mixed effect models revealed that the presence (or absence) of variants A3, B3 and C3 in a cow's genotype was associated with the quantity of some milk FA methyl esters (FAMEs), with different variants having different effects as detailed in Table 1. The presence of variant A3 (the most common variant) was associated with decreased C15:1, C18:1 trans-11, C18:1 all trans, C18:2 trans-9, cis-12, C22:0 and C24:0 levels but increased levels of C12:1 and C13:0 iso (p<0.05). Variant B3 was revealed to be associated with reduced levels of C6:0, C8:0, C11:0, C13:0 and C20:0 but increased C17:0 iso and C24:0 levels (p<0.05). Variant C3 was associated with decreased C17:0 iso level but an increased level of C20:0 (p<0.05).
Table 1Associations between bovine leptin gene variants with average quantity of individual and grouped milk fatty acid methyl ester (FAME) in New Zealand (NZ) HF × J cows.
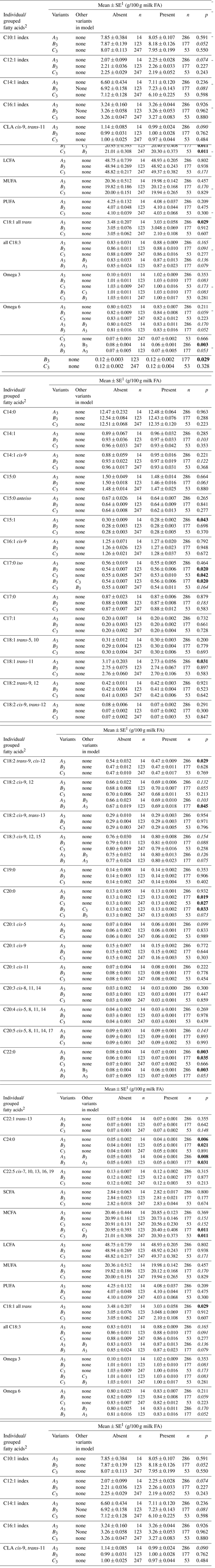
1 Predicted means and standard error of those means derived from general linear mixed-effects models (GLMMs). Cow age (categorical variable), LEP variants (categorical variable), herd (categorical variable) and days in milk (continuous variable) were fitted to the model as fixed effects. 0.05<p<0.2 in italics, p<0.05 in bold. 2 SCFA – short-chain fatty acid; MCFA – medium-chain fatty acid; LCFA – long-chain fatty acid; MUFA – monounsaturated fatty acid; PUFA – polyunsaturated fatty acid; UFA – unsaturated fatty acid; SFA – saturated fatty acid. The unit (g/100 g milk FA) applied to all FAs except for the FA indices which had a unit of %.
3.3 Genotype models
Only the genotypes A3A3 (n = 70), A3B3 (n = 166) and A3C3 (n = 50) with a frequency greater than 5 % were analysed. The other genotypes B3B3 (n = 11) and C3C3 (n = 3) were not included in this model. The composition of milk fat was affected by genotype, and the results were consistent with the findings of the variant presence/absence models. Cows carrying the A3A3 (most common) genotype contained higher levels of saturated fatty acids (SFAs), but when one copy of the A3 variant was replaced by B3 or C3, the resulting heterozygous genotype (A3B3 or A3C3) was associated with changed levels of SFAs in milk. Cows carrying the A3B3 genotype was associated with increased levels of C22:0 and C24:0 but decreased C8:0, C10:0, C11:0, C13:0, C15:0 and grouped MCFA levels (p<0.05). A3C3 was found to be associated with decreased levels of C10:0, C11:0, C13:0 and grouped MCFA (p<0.05; Table 2).
Table 2Associations between milk fatty acid levels and leptin genotypes.
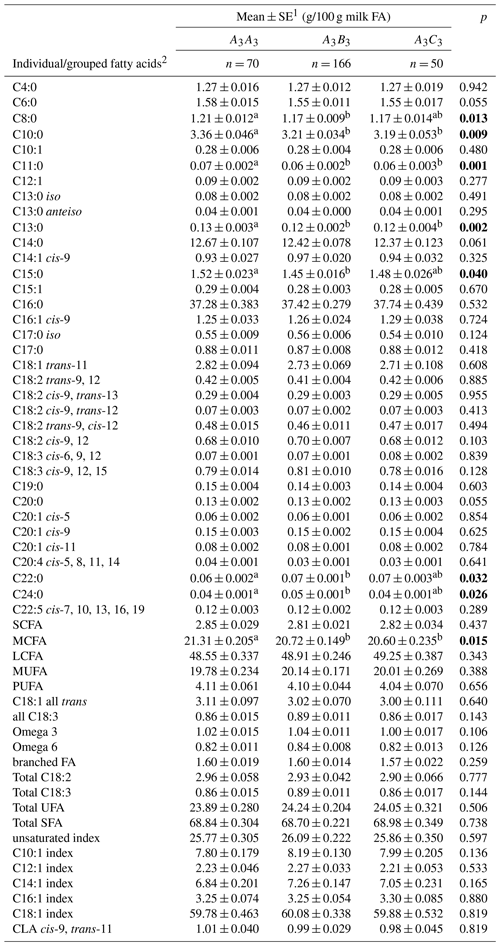
1 Predicted means and standard error of those means derived from general linear mixed-effects models (GLMMs). Cow age (categorical variable), leptin genotypes (categorical variable), herd (categorical variable) and days in milk (continuous variable) were fitted to the model as fixed effects. Means within a row that do not share a superscript letter (a or b) are separated by Bonferroni test at p<0.05. 0.05<p<0.02 in italics, while p<0.05 is in bold. 2 SCFA – short-chain fatty acid; MCFA – medium-chain fatty acid; LCFA – long-chain fatty acid; MUFA – monounsaturated fatty acid; PUFA – polyunsaturated fatty acid; UFA – unsaturated fatty acid; SFA – saturated fatty acid. The unit (g/100 g milk FA) applied to all FAs except for the FA indices which had a unit of %.
This is the first study investigating the effect of leptin gene variations in exon 3 with composition of milk FA in NZ HF × J cows farmed wholly outdoors on pasture.
Overall, the results presented here revealed associations between variation in the leptin gene and the composition of milk fat. Cows carrying the A3A3 genotype had higher levels of SFAs, but when one copy of the A3 variant is replaced by a B3 variant, the resulting heterozygous genotype A3B3 had decreased levels of SFA.
In an analysis of the effect of LEP nucleotide sequence variation on the FA profile of cattle muscle fat, Orrù et al. (2011) investigated the effect of c.239C/T (p.Ala80Val – also identified in this study) in 103 Simmental bulls. They revealed that the C allele (the allele with alanine at amino acid 80 – equivalent to the A3 variant here) was associated with increased meat C14:1 and C14 index. In contrast, our study revealed the presence of variant A3 was associated with increased C12:1 but decreased C15:1 and C18:1 all trans. In addition, the B3 variant identified in this study, which carries the T in the nucleotide substitution c.239C/T (has a valine residue at position 80), was associated with a decrease in some short- and medium-chain SFAs. Taken together, the observation that the C and T alleles of c.239C/T appeared to affect the composition of FAs in meat and milk differently suggests further investigation of this substitution and its effects is required.
In another study (Avondo et al., 2019), the effects of variation in a LEP intron 1 microsatellite sequence and its interaction with milk FA composition, diet, milk traits, and metabolic state in Girgentana lactating goats at mid-lactation were investigated. It was revealed that the composition of milk FA was strongly influenced by LEP genotype. Goats with the homozygous genotype 266 bp/266 bp (L genotype) had lower levels of SFA but increased levels of MUFA and PUFA, compared to goats with the heterozygous genotype 266 bp/264 bp (H genotype). Although our results also showed a decrease in the levels of SFA, it is difficult to specifically link our results to the work of Avondo et al. (2019) because of the differences in the gene regions studied and the species investigated. In the Avondo et al. (2019) study, the differences described between the LEP genotypes suggested that the L genotype could be associated with a higher utilization of body fat reserves. This is consistent with the finding of higher levels of MUFA and PUFA and lower levels of SFA found with the increased mobilization of FAs from adipose tissue in other studies (Palmquist et al., 1993; Vrankovic et al., 2017). It may also be consistent with the hypothesis of increased demand for energy as reported by Di Gregorio et al. (2014) for the L genotype.
The leptin gene from both cattle and goats map to chromosome 4, and on that chromosome there are quantitative trait loci (QTLs) for fat yield and percentage in milk (Cattle QTL database https://www.animalgenome.org/cgi-in/QTLdb/BT/index, last access: 10 July 2020) and FA composition (Li et al., 2014). This suggests it would be worthwhile undertaking further research into the role of bovine LEP and variation in the gene in the mobilization and utilization of body fat reserves.
These previous reports, along with the findings we report, appear to contradict the findings of Marchitelli et al. (2013). Their study did not reveal any association between the p.Arg25Cys SNP in LEP exon 2 and milk FA traits in Jersey, Piedmontese and Valdostana cattle breeds. A number of factors may have been responsible for this disparity in findings, including the obvious difference in gene region examined and the potential effect of breed differences. While Marchitelli et al. (2013) investigated the effect of the exon 2 region carrying the non-synonymous p.Arg25Cys SNP on milk FA traits, our study examined the effect of exon 3 carrying the non-synonymous p.Ala80Val SNP. Even though both nucleotide sequence variations are non-synonymous, it is likely that these SNPs will affect the concentration of milk FAs differently, since they are located on different parts of the gene. Also, while we investigated 300 NZ cross-bred HF × J cows (albeit of no fixed breed proportion), Marchitelli et al. (2013) investigated 90 cows in total which included the Italian Piedmontese, Valdostana and Jersey breeds. These breeds differ in terms of milk-related traits, especially in the composition of milk FAs. For example, milk from Jersey cows contains higher concentrations of some short- and medium-chain SFA but lower concentrations of some UFA (Arnould and Soyeurt, 2009). Other studies have also suggested that breed is an important factor that affects milk FA content (Karijord et al., 1982; Lawless et al., 1999). It therefore seems plausible that differences in breed may underlie the discrepancies in findings.
Another possible reason for the differences in findings can be attributed to diet. In our investigation, the NZ HF × J dairy cows were all grazed on pasture (a mixture of perennial ryegrass and white clover), whereas the cows chosen by Marchitelli et al. (2013) were fed with “unifeed” (corn silage and concentrates). The pasture-based production system increases the amount of PUFA and conjugated linoleic acids (CLAs) in the milk as suggested by Chilliard et al. (2001) and Dewhurst et al. (2006). In this context, differences in diet may have contributed to the disparity between our findings and those of Marchitelli et al. (2013), especially considering a previous report that suggested diet may affect the production of milk fat (Stelwagen, 2011).
The findings here suggest that cows carrying the variant leptin genotype A3B3 (where the B3 variant in exon 3 with accession number MN119554 carries the p.Ala80Val SNP) are associated with decreased SFA levels in milk. Since heterozygous cows A3B3 had reduced SFA levels, cows with the B3B3 genotype might therefore have much lower levels of SFA in their milk. Unfortunately, since there were insufficient cattle with the homozygous genotypes B3B3 in the cattle investigated, further studies involving larger sample sizes across different farms and breeds are needed to validate this claim.
The original data are available upon request to the corresponding author.
ILH carried out the experiment and the statistical analysis and wrote the paper. JGHH and HZ designed and supervised the experiment, helped with interpretation of the results, and edited the paper. All authors reviewed and approved the final paper.
The authors declare that they have no conflict of interest.
The authors will like to thank the staff of the Gene-Marker Laboratory at Lincoln University for technical support and facilities provided.
This paper was edited by Steffen Maak and reviewed by Marcella Avondo and Ankit Magotra.
Arnould, V. M. R., and Soyeurt, H.: Genetic variability of milk fatty acids, J. Appl. Genet., 50, 29–39, https://doi.org/10.1007/BF03195649, 2009.
Avondo, M., Trana, A. D., Valenti, B., Criscione, A., Bordonaro, S., Angelis, A. D., Giorgio, D., and Gregorio, P. D.: Leptin Gene Polymorphism in Goats Fed with Diet at Different Energy Levels: Effects on Feed Intake, Milk Traits, Milk Fatty Acids Composition, and Metabolic State, Anim., 9, 424, https://doi.org/10.3390/ani9070424, 2019.
Byun, S. O., Fang, Q., Zhou, H., and Hickford, J. G. H.: An effective method for silver-staining DNA in large numbers of polyacrylamide gels, Anal. Biochem., 385, 174–175, https://doi.org/10.1016/j.ab.2008.10.024, 2009.
Chilliard, Y., Ferlay, A., and Doreau, M.: Effect of different types of forages, animal fat or marine oils in cow's diet on milk fat secretion and composition, especially conjugated linoleic acid (CLA) and polyunsaturated fatty acids, Livest. Prod. Sci., 70, 31–48, https://doi.org/10.1016/S0301-6226(01)00196-8, 2001.
Dewhurst, R. J., Shingfield, K. J., Lee, M. R. F., and Scollan, N. D.: Increasing the concentrations of beneficial polyunsaturated fatty acids in milk produced by dairy cows in high-forage systems. Anim. Feed Sci. Technol., 131, 168–206, https://doi.org/10.1016/j.anifeedsci.2006.04.016, 2006.
De Matteis, G., Scatà, M. C., Grandoni, F., Petrera, F., Abeni, F., Catillo, G., Napolitano, F., and Moioli, B.: Association analyses of single nucleotide polymorphisms in the leptin and leptin receptor genes on milk and morphological traits in Holstein cows, Open J. Anim. Sci., 2, 174–182, https://doi.org/10.4236/ojas.2012.23024, 2012.
Di Gregorio, P., Di Trana, A., Celi, P., Claps, S., and Rando, A.: Comparison of goat, sheep, cattle and water buffalo leptin (LEP) genes and effects of the Intron 1 microsatellite polymorphism in goats, Anim. Prod. Sci., 54, 1258–1262, https://doi.org/10.1071/AN14101, 2014.
Feuermann, Y., Mabjeesh, S. J., and Shamay, A.: Leptin affects prolactin action on milk protein and fat synthesis in the bovine mammary gland, J. Dairy Sci., 87, 2941–2946, https://doi.org/10.3168/jds.S0022-0302(04)73425-6, 2004.
Giblin, L., Butler, S. T., Kearney, B. M., Waters, S. M., Callanan, M. J., and Berry, D. P.: Association of bovine leptin polymorphisms with energy output and energy storage traits in progeny tested Holstein-Friesian dairy cattle sires, BMC Genet., 11, 73, https://bmcgenet.biomedcentral.com/articles/10.1186/1471-2156-11-73 (last access: 17 March 2020), 2010.
Gong, H., Zhou, H., and Hickford, J. G. H.: Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep, Mol. Biol. Rep., 38, 31–35, https://doi.org/10.1007/s11033-010-0074-6, 2011.
Hajihosseinlo, A., Hashemi, A., and Sadeghi, S.: Association between polymorphism in exon 3 of leptin gene and growth traits in the Makooei sheep of Iran, Livest. Res. Rural Dev., 24, 543–546, 2012.
Halaas, J. L., Gajiwala, K. S., Maffei, M., Cohen, S. L., Chait, B. T., Rabinowitz, D., Lallone, R. L., Burley, S. K., and Friedman, J. M.: Weight-reducing effects of the plasma protein encoded by the obese gene, Science, 269, 543–546, https://doi.org/10.1126/science.7624777, 1995.
Haruna, I. L., Hadebe, S. A., Oladosu, O. J., Mahmoud, G., Zhou, H., and Hickford, G. H. J.: Identification of novel nucleotide sequence variations in an extended region of the bovine leptin gene (LEP) across a variety of cattle breeds from New Zealand and Nigeria, Arch. Anim. Breed., 63, 241–248, https://doi.org/10.5194/aab-63-241-2020, 2020.
Houseknecht, K. L., Baile, C. A., Matteri, R. L., and Spurlock, M. E.: The biology of leptin: A review, J. Anim. Sci., 76, 1405–1420, https://doi.org/10.2527/1998.7651405x, 1998.
Karijord, O., Standal, N., and Syrstad, O.: Sources of variation in composition of milk fat, Z. Tierz. Zuchtungsbio., 99, 81–93, 1982.
Kawaguchi, F., Okura, K., Oyama, K., Mannen, H., and Sasazaki, S.: Identification of leptin gene polymorphisms associated with carcass traits and fatty acid composition in Japanese Black cattle, Anim. Sci. J., 88, 433–438, https://doi.org/10.1111/asj.12672, 2017.
Lagonigro, R., Wiener, P., Pilla, F., Woolliams, J. A., and Williams, J. L.: Short Communication: A new mutation in the coding region of the bovine leptin gene associated with feed intake, Anim. Genet., 34, 371–374, https://doi.org/10.1046/j.1365-2052.2003.01028.x, 2003.
Lawless, F., Stanton, C., Escop, P., Devery, R., Dillon, P., and Murphy, J. J.: Influence of breed on bovine milk cis-9, trans-11-conjugated linoleic acid content, Livest. Prod. Sci., 62, 43–49, https://doi.org/10.1016/S0301-6226(99)00053-6, 1999.
Li, C., Sun, D., Zhang, S., Wang, S., Wu, X., Zhang, Q., Liu, L., Li, Y., and Qiao, L.: Genome Wide Association Study Identifies 20 Novel Promising Genes Associated with Milk Fatty Acid Traits in Chinese Holstein, PLoS ONE, 9, e96186, https://doi.org/10.1371/journal.pone.0096186, 2014.
Li, Y., Zhou, H., Cheng, L., Edwards, G. R., and Hickford, J. G. H.: Effect of DGAT1 variant (K232A) on milk traits and milk fat composition in outdoor pasture-grazed dairy cattle, New Zeal. J. Agr. Res., 64, 101–113, https://doi.org/10.1080/00288233.2019.1589537, 2019.
Liefers, S. C., te Pas, M. F. W., Veerkamp, R. F., and van der Lende, T.: Associations between leptin gene polymorphisms and production, live weight, energy balance, feed intake, and fertility in Holstein heifers, J. Dairy Sci., 85, 1633–1638, https://doi.org/10.3168/jds.S0022-0302(02)74235-5, 2002.
Marchitelli, C., Contarini, G., De Matteis, G., Crisa, A., Pariset, L., Scata, M. C., Catillo, G., Napolitano, F., and Moioli, B.: Milk fatty acid variability: Effect of some candidate genes involved in lipid synthesis, J. Dairy Res., 80, 165–173, https://doi.org/10.1017/S002202991300006X, 2013.
Orrù, L., Cifuni, G. F., Piasentier, E., Corazzin, M., Bovolenta, S., and Moioli, B.: Association analyses of single nucleotide polymorphisms in the LEP and SCDI genes on the fatty acid profile of muscle fat in Simmental bulls, Meat Sci., 87, 344–348, https://doi.org/10.1016/j.meatsci.2010.11.009, 2011.
Palmquist, D. L., Beaulieu, A. D., and Barbano, D. M.: Feed and Animal Factors Influencing Milk Fat Composition, J. Dairy Sci., 76, 1753–1771, https://doi.org/10.3168/jds.S0022-0302(93)77508-6, 1993.
Pegolo, S., Cecchinato, A., Mele, M., Conte, G., Schiavon, S., and Bittante, G.: Effect of candidate gene polymorphisms on the detailed fatty acids profile determined by gas chromatography in bovine milk, J. Dairy Sci., 99, 4558–4573, https://doi.org/10.3168/jds.2015-10420, 2016.
Pomp, D., Zou, T., Clutter, A. C., and Barendse, W.: Rapid communication: mapping of leptin to bovine chromosome 4 by linkage analysis of a PCR-based polymorphism, J. Anim. Sci., 75, 14–27, https://doi.org/10.2527/1997.7551427x, 1997.
Stelwagen, K.: Mammary Gland, Milk Biosynthesis and Secretion Lactose, in: Encyclopedia of Dairy Science, edited by: Fuquay, J. W., San Diego, Academic Press, 367–372, 2016.
Tang-Christensen, M., Havel, P. J, Jacobs, R. R., Larsen, P. J., and Cameron, J. L.: Central administration of leptin inhibits food intake and activates the sympathetic nervous system in rhesus macaques, J. Clin. Endocrinol. Metab., 84, 711–717, https://doi.org/10.1210/jcem.84.2.5458, 1999.
Vrankovic, L., Aladrovic, J., Octenjak, D., Bijelic, D., Cvetnic, L., and Stojevic, Z.: Milk fatty acid composition as an indicator of energy status in Holstein dairy cows, Arch. Anim. Breed., 60, 205–212, https://doi.org/10.5194/aab-60-205-2017, 2017.
Zhou, H., Hickford, J. G. H., and Fang, Q.: A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification, Anal. Biochem., 354, 159–161, https://doi.org/10.1016/j.ab.2006.03.042, 2006.