the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Variation in KRTAP6-1 affects wool fibre diameter in New Zealand Romney ewes
Wenhao Li
Hua Gong
Huitong Zhou
Jiqing Wang
Shaobin Li
Xiu Liu
Yuzhu Luo
Jon G. H. Hickford
Variation in KRTAP6-1 has been reported to affect wool fibre traits in Merino cross-breed sheep and Chinese Tan sheep, but little is known about whether these effects persist in other breeds. In this study, variation in KRTAP6-1 was investigated in 290 New Zealand (NZ) Romney ewes sired by 16 different rams. Polymerase chain reaction single-stranded conformational polymorphism (PCR-SSCP) analysis revealed four variants (A, B, E and F) of KRTAP6-1, and nine genotypes (AA, AB, AE, AF, BB, BE, BF, EE and FF) in these ewes. Among the 243 ewes that had genotypes with a frequency of over 5 % (i.e. AA, AB and BB), the presence of A was found to be associated with reduced mean fibre diameter (MFD) and increased coefficient of variation in fibre diameter (CVFD), whereas the presence of B had a trend of association with decreased coarse edge measurement (CEM). A genotype effect was also detected for MFD and CVFD. No associations were detected for fibre diameter standard deviation (FDSD), mean fibre curvature (MFC) and medulation. These results suggest that variation in KRTAP6-1 affects wool fibre diameter in NZ Romney ewes, confirming the finding in Merino cross-breed sheep.
- Article
(1580 KB) - Full-text XML
- BibTeX
- EndNote
Keratin-associated proteins and keratin intermediate filament proteins are the major structural proteins of wool, and they cross-link to form the skeleton of wool fibres (Powell and Rogers, 1997). The keratin-associated proteins (KAPs) are classified into three groups according to their amino acid composition: the high sulfur family (HS; ≤30 mol % cysteine), the ultra-high sulfur family (UHS; >30 mol % cysteine) and the high glycine and tyrosine family (HGT; 35–60 mol % glycine and tyrosine) (Gong et al., 2016). Of these, the HGT-KAPs are the first group of KAPs to be produced in active wool follicles, and soon after the synthesis of the keratins (Rogers, 2006). They vary considerably in abundance between and within sheep breeds (Gillespie, 1990).
The KAP6 family is a diverse family from the HGT-KAP group. The family has five gene members (KRTAP6-1, KRTAP6-2, KRTAP6-3, KRTAP6-4 and KRTAP6-5) in sheep (Zhou et al., 2016), and they are located on chromosome 1. They are clustered with six other HGT-KAP genes (KRTAP7-1, KRTAP8-1, KRTAP8-2, KRTAP20-1, KRTAP20-2 and KRTAP22-1) (Bai et al., 2018; Gong et al., 2014, 2019; Li et al., 2017a) and six HS-KAP genes (KRTAP11-1, KRTAP13-1, KRTAP15-1, KRTAP24-1, KRTAP26-1 and KRTAP28-1) (Bai et al., 2019; Gong et al., 2011a, 2012b; Wang et al., 2017; Zhou et al., 2012). Five variant sequences have been identified at the KRTAP6-1 locus and this variation has been reported to be associated with various mean fibre diameter (MFD)-associated traits in Merino cross-breed sheep (Zhou et al., 2015), and fibre length and crimping in early life in Chinese Tan sheep (Tao et al., 2017). These findings suggest variation in KRTAP6-1 affects wool traits, but little is known about whether the effect is preserved across other breeds, including breeds that produce strong (or high MFD) wool.
The New Zealand (NZ) Romney sheep is a dual-purpose breed for meat and wool production. It is the most common sheep breed in NZ and accounts for over 47 % of the 27.4 million sheep wintered-over in June 2017 (Beef and Lamb New Zealand, 2018). The NZ Romney wool typically has a MFD of over 36 µm.
In this study, variation in KRTAP6-1 was investigated in NZ Romney ewes using Polymerase chain reaction single-stranded conformational polymorphism (PCR-SSCP) analysis, and its association with six different wool traits was analysed to determine whether the KRTAP6-1 variation affected these traits in this strong-wool breed.
This study was carried out in accordance with the Animal Welfare Act 1999 (NZ Government), and the collection of sheep blood drops by nicking sheep ears is covered by Sect. 7.5, Animal Identification, of the Sheep and Beef Cattle Code of Welfare 2010 (NZ Government).
2.1 Sheep blood and wool sample collection and preparation
Two hundred and ninety NZ Romney ewes from 16 different sires sourced from seven farms in New Zealand were investigated initially. A blood sample from each ewe was collected onto TFN paper (Munktell Filter AB, Sweden) and genomic DNA was purified from dried blood spots using a two-step purification method described in Zhou et al. (2006).
A wool sample from the mid-side region of each ewe was collected at “hogget shearing”, with ages ranging from 15 to 17 months old, depending on the farm. These samples were measured for mean fibre diameter (MFD), fibre diameter standard deviation (FDSD), coefficient of variation in fibre diameter (CVFD), coarse edge measurement (CEM; the percentage of fibres that are 10 µm greater than the mean fibre diameter), mean fibre curvature (MFC), and medulation. The measurements of these wool traits were undertaken by Pastoral Measurements Limited (Timaru, NZ).
2.2 Genotyping of ovine KRTAP6-1
A fragment of KRTAP6-1 containing the entire coding region was amplified using the PCR primers described in Zhou et al. (2015). These primers were 5′-TCTACCCGAGAACAACCTC-3′ (forward) and 5′-AGGCAAGTCTTTAGTAGGAC-3′ (reverse), and they were synthesized by Integrated DNA Technologies (Coralville, IA, USA). PCR amplification was performed using a 15 µL reaction consisting of a 1.2 mm punch of genomic DNA from the TFN card, 0.25 µM of each primer, 150 µM of each deoxynucleoside triphosphate (dNTP) (Bioline, London, UK), 2.5 mM Mg2+, 0.5 U of Taq DNA polymerase (Qiagen, Hilden, Germany) and 1× the reaction buffer supplied with the enzyme. The thermal profile included an initial denaturation at 94 ∘C for 2 min, followed by 35 cycles of 94 ∘C for 30 s, 59 ∘C for 30 s and 72 ∘C for 30 s, and a final extension step at 72 ∘C for 5 min.
The PCR amplicons were subject to single-stranded conformational polymorphism (SSCP) analysis to differentiate variants. A 1 µL aliquot of each PCR amplicon was mixed with 7 µL of loading dye (98 % formamide, 10 mM EDTA, 0.025 % bromophenol blue, 0.025 % xylene cyanol) and denatured at 90 ∘C for 5 min. The sample was rapidly cooled on wet ice and then loaded onto a 16 cm×18 cm, 12 % polyacrylamide (acrylamide–bisacrylamide 37.5:1) gel containing 1 % glycerol. Electrophoresis was carried out in 0.5× TBE buffer at 330 V and 12 ∘C for 16 h, and the gel was silver stained using the method by Byun et al. (2009).
2.3 Sequencing of variants and sequence analysis
Polymerase chain reaction amplicons representing different SSCP banding patterns from sheep that appeared to be homozygous were sequenced in both directions at the Lincoln University DNA sequencing facility (Lincoln University, New Zealand). Variants that were only found in heterozygous ewes were sequenced using an approach described by Gong et al. (2011b). Briefly, a band corresponding to the variant was removed as a slice from the polyacrylamide gel, mashed-up and then used as a template for re-amplification with the original primers. This second amplicon was then sequenced directly.
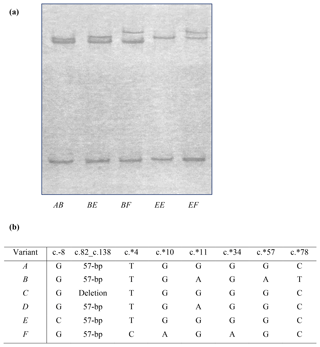
Figure 1Polymorphism of ovine KRTAP6-1. (a) Four variants (A, B, E and F) of ovine KRTAP6-1 were detected in New Zealand Romney ewes by PCR single-stranded conformational polymorphism analysis. (b) Sequence comparison of the four ovine KRTAP6-1 variants identified in this study, and those previously identified variants (C and D), revealed three new single-nucleotide polymorphisms (SNPs) at c.∗4, c.∗10 and c.∗34.
DNA sequence alignments, and translation to putative amino acid sequences (this assumes the gene is expressed) were carried out using DNAMAN (version 5.2.10, Lynnon BioSoft, Vaudreuil, Quebec, Canada).
2.4 Statistical analyses
Statistical analysis was performed using Minitab version 16 (Minitab Inc., State College, PA, USA). Association analyses were only carried out for the ewes that had variants and genotypes with frequencies of over 5 % (and therefore with sufficient sample size) and the ewes with other genotypes were removed from the association study.
General linear mixed models (GLMMs) were used to assess the effect of the absence or presence of individual variants on the wool traits, and also compare various wool traits between ewes with different genotypes. Bonferroni corrections were applied to these genotype models to reduce the chance of obtaining false positive results during the multiple comparisons. Sire was incorporated as random factors in the models. As each farm had different sires, the ewes from different farms were therefore from different sires; accordingly, the wool samples from different sires were taken at different ages. This meant that sire, farm and ewe age were confounded. Sire had the greatest effect on the wool traits, and therefore only sire was included in the models. All wool samples were collected from ewes, hence gender was not fitted into the models.
3.1 Polymorphism of KRTAP6-1 in NZ Romney ewes
Four unique PCR-SSCP patterns were detected among the NZ Romney ewes (Fig. 1a). Sequencing of amplicons representative of these banding patterns revealed three previously identified KRTAP6-1 variants A, B and E (Zhou et al., 2015; Tao et al., 2017) and one newly identified variant named variant F. The detection of variant F resulted in the definition of three new single-nucleotide polymorphisms (SNPs) at c.∗4, c.∗10 and c.∗34, and these SNPs were in linkage (Fig. 1b). Two previously identified variants, C and D (Zhou et al., 2015), were not found in these NZ Romney ewes.
Nine genotypes were observed, with the genotypes and frequencies being AA (17.5 %), AB (43.9 %), AE (2.6 %), AF (3.4 %), BB (22.5%), BE (3.9 %), BF (4.7 %), EE (0.5 %) and FF (1.0 %). These gave varying frequencies of 42.4 %, 48.7 %, 3.8 % and 5.1 % for A, B, E and F, respectively.
Table 1Association between the presence or absence of ovine KRTAP6-1 variants and wool traits in New Zealand Romney ewes.
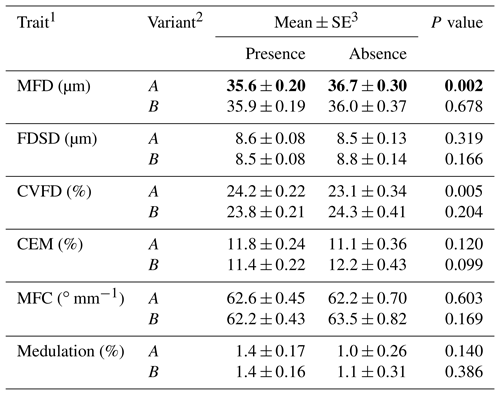
1 MFD – mean fibre diameter; FDSD – fibre diameter standard deviation; CVFD – coefficient of variation in fibre diameter; CEM – coarse edge measurement (the percentage of fibres that are 10 µm greater than the mean fibre diameter); MFC – mean fibre curvature. 2 Of 243 Romney ewes that had common genotypes (i.e. AA, AB or BB), A was present in 179 ewes and absent in 64 ewes, and B was present in 192 ewes and absent in 51 ewes. 3 Estimated marginal means and standard errors (SE) of those means from general linear mixed models, with P<0.05 being shown in bold.
Table 2Association of ovine KRTAP6-1 genotypes and wool traits in New Zealand Romney ewes.
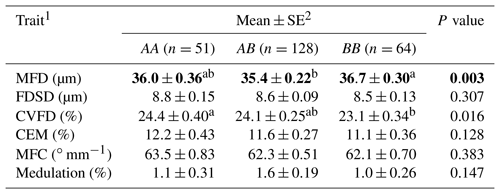
1 MFD – mean fibre diameter; FDSD – fibre diameter standard deviation; CVFD – coefficient of variation in fibre diameter; CEM – coarse edge measurement (the percentage of fibres that are 10 µm greater than the mean fibre diameter); MFC – mean fibre curvature. 2 Estimated marginal means and standard errors (SE) of those means from general linear mixed models. Means within rows that do not share a superscript letter are different at P<0.05, and are shown in bold.
3.2 Association between KRTAP6-1 variation and wool traits
Of the 243 ewes that had the common genotypes AA, AB and BB (occurring at frequencies of over 5 %), the presence of variant A was found to be associated with a decrease in MFD and an increase in CVFD and the presence of B was found to have a trend of association with decreased CEM (Table 1). Wool from the AA ewes had a higher CVFD than BB ewes, with AB ewes being intermediate (Table 2). Wool produced by BB ewes had higher MFD than AB ewes but was not different to AA ewes (Table 2). No associations were detected for FDSD, MFC or medulation.
This study describes genetic variation in ovine KRTAP6-1 in 15–17-month-old NZ Romney ewes and its association with MFD-associated wool traits in this strong-wool breed. At this age, wool measurement is important, as in most NZ Romney sheep production systems; this is when ewe selection decisions are commonly made and the ewes kept after selection are mated for the first time, entering the main ewe flock.
The detection of three previously identified and one newly identified variant, in these NZ Romney ewes, suggests KRTAP6-1 exhibits a moderate level of variation in this breed but the nature of the variation in this breed appears to be different to that reported in Merino cross-breed sheep (Zhou et al., 2015) and Chinese Tan sheep (Tao et al., 2017). In particular, the C variant that contains a 57 bp deletion, and that was found to be associated with higher MFD, FDSD and CVFD in Merino cross-breed sheep (Zhou et al., 2015) was not found in these NZ Romney ewes. Variant D that occurred at a frequency of over 10 % in Chinese Tan sheep, and was found to be associated with increased straightened fibre length at birth (Tao et al., 2017), was also not found in the NZ Romney ewes. Instead a new variant (F) was detected.
The detection of a new variant brings the number of ovine KRTAP6-1 variants from five to six, and the number of SNPs from four to seven in a 343 bp fragment of the gene (excluding the primer binding regions). This gives a density of over 20 SNPs per kilobase pair, which is much higher than the average density of 4.9 SNPs per kilobase pair across the sheep genome (Kijas et al., 2009). What is more, it might be that more variants are identified when more sheep from more breeds are investigated. These SNPs, together with the presence of 57 bp indel (Zhou et al., 2015) suggest that ovine KRTAP6-1 is a polymorphic gene.
Sequence variation has been reported in many other ovine KRTAP genes (Gong et al., 2012a, 2019; Li et al., 2017a, b, 2018), but the sequence variation identified in KRTAP6-1 has some unique features. First is the location of the sequence variation. All of the SNPs identified in ovine KRTAP6-1 are located either upstream or downstream of the coding region, but not in the coding region itself. The absence of coding region SNPs has not been reported for any of the other ovine KRTAP genes identified (Gong et al., 2016, 2019; Li et al., 2017a, b, 2018), except KRTAP8-2, which has only one SNP at a position 21 bp upstream of the TATA box (Gong et al., 2014). The unusual location of SNPs suggests that ovine KRTAP6-1 may be constrained as regards variation in its protein structure and/or function, but possibly exhibit flexibility in its level of expression.
The second notable feature is the linkage of the SNPs. For example, linkage is observed for the variation at c.∗4, c.∗10 and c.∗34 and further linkage is detected for the variations at c.∗57 and c.∗78. The linkage could suggest that gene conversion may have occurred at a location downstream of the coding region and that this may have led to the generation of these SNPs. Gene conversion has been described for HS-KRTAP1-n (Rogers et al., 1994; Zhou et al., 2019); for the KRTAP1-n gene, conversion is observed within the coding region and appears to be the mechanism responsible for the concerted evolution of the KRTAP1-n coding region (Zhou et al., 2019).
The detection of variant and genotype associations with MFD and CVFD, but not with FDSD, suggests that variation in KRTAP6-1 affects the average of the fibre diameters, but not the distribution of individual fibre diameters around that average. This is because %, and hence a decrease in MFD would lead to an increase in CVFD, if FDSD is unchanged. The failure to detect a difference in MFD for genotype AA compared to AB and BB, a situation that seems rather odd, may be due to a number of factors. Firstly, it may be an effect of sample size, as genotype AA was only found in 51 ewes, which is the smallest group among the genotypes used for the association analyses. Second, may be the level of variation observed for the wool traits. Genotype AA ewes had higher standard errors (SEs) for the adjusted means for all the wool traits compared to AB or BB ewes. High SEs may suggest higher levels of variation in traits, and suggesting that some uncontrolled variable is unaccounted for. For example, variant A, as described in this breed, might actually represent more than one form of the gene, with the difference between those forms occurring outside of the amplified region. Given that variation in KRTAP6-1 described here is located outside of the coding region, it remains possible that even more variation of consequence to the activity of the gene occurs in regions further upstream or downstream. It is also possible that variant A may be linked to variation in other proximal KRTAP genes, and consequently there is some kind of haplotype effect. This would once again require further investigation.
The effect of the KRTAP6-1 variation detected in the NZ Romney ewes appears to be different to that detected in Merino cross-breed sheep, in which FDSD and prickle factor (comparable to CEM described for coarse wool) are also affected (Zhou et al., 2015). This suggests that the role that KRTAP6-1 plays in regulating wool fibre traits may differ in fine- and strong-wool breeds. Zhang et al. (2017) reported that KRTAP6-1 was differentially expressed in Chinese Merino (fine wool) and Small Tailed Han (strong wool) sheep, with a higher level of expression in the Merino sheep compared to the Small Tailed Han sheep. Reduced expression of KRTAP6-1 may lead to a reduced effect on wool traits, which appears to be consistent with the results obtained in NZ Romney ewes. This would require further investigation; nevertheless, the results from this study demonstrate that further sequence variation occurs in ovine KRTAP6-1, and that this sequence variation also affects wool fibre diameter traits.
The original data are available upon request to the corresponding authors.
HG, HZ, YL and JGHH conceived and designed the experiments. WL and HG performed the experiments. WL, HZ, HG, JW, SL, XL and JGHH analysed the data. WL, HZ and JGHH wrote the article with contributions from other co-authors. All authors reviewed and commented on the article.
The authors declare that they have no conflict of interest.
The authors thank Qian Fang and Andrea Hogan for technical assistance.
This research has been supported by the fund of the College of Animal Science and Technology, Gansu Agricultural University (grant no. XMXTSXK-28).
This paper was edited by Steffen Maak and reviewed by Türker Savaş and one anonymous referee.
Bai, L., Gong, H., Zhou, H. , Tao, J., and Hickford, J. G. H.: A nucleotide substitution in the ovine KAP20-2 gene leads to a premature stop codon that affects wool fibre curvature, Anim. Genet., 49, 357–358, https://doi.org/10.1111/age.12668, 2018.
Bai, L., Wang, J., Zhou, H., Gong, H., Tao, J., and Hickford, J. G. H.: Identification of ovine KRTAP28-1 and its association with wool weight and mean fibre diameter-associated traits, Animals, 9, 142, https://doi.org/10.3390/ani9040142, 2019.
Beef and Lamb New Zealand: Compendium of Farm Facts of New Zealand, 42 Edn., Publication No. P18010, Beef + Lamb New Zealand, Economic Service, available at: https://beeflambnz.com/knowledge-hub/PDF/compendium-farm-facts, 2018.
Byun, S. O., Fang, Q., Zhou, H., and Hickford, J. G. H.: An effective method for silver-staining DNA in large numbers of polyacrylamide gels, Anal. Biochem., 385, 174–175, https://doi.org/10.1016/j.ab.2008.10.024, 2009.
Gillespie, J. M.: The proteins of hair and other hard a-keratins, in: Cellular and Molecular Biology of Intermediate Filaments, edited by: Goldman, R. D. and Steinert, P. M., Plenum Press, New York, 95–128, 1990.
Gong, H., Zhou, H., Dyer, J. M., Plowman, J. E., and Hickford, J. G. H.: Identification of the keratin-associated protein 13-3 (KAP13-3) gene in sheep, Open J. Genet., 1, 60–64, https://doi.org/10.4236/ojgen.2011.13011, 2011a.
Gong, H., Zhou, H., and Hickford, J. G. H.: Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep, Mol. Biol. Rep., 38, 31–35, https://doi.org/10.1007/s11033-010-0074-6, 2011b.
Gong, H., Zhou, H., McKenzie, G. W., Yu, Z., Clerens, S., Dyer, J. M., Plowman, J. E., Wright, M. W., Arora, R., Bawden, C. S., Chen, Y., Li, J., and Hickford, J. G. H.: An updated nomenclature for keratin-associated proteins (KAPs), Int. J. Biol. Sci., 8, 258–64, https://doi.org/10.7150/ijbs.3278, 2012a.
Gong, H., Zhou, H., Dyer, J. M., and Hickford, J. G. H.: Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep., 38, 5429–5433, https://doi.org/10.1007/s11033-011-0697-2, 2012b.
Gong, H., Zhou, H., Dyer, J., and Hickford, J. G. H.: The sheep KAP8-2 gene, a new KAP8 family member that is absent in humans, Springer Plus, 3, 528, https://doi.org/10.1186/2193-1801-3-528, 2014.
Gong, H., Zhou, H., Forrest, R. H., Li, S., Wang, J., Dyer, J. M., Luo, Y., and Hickford, J. G. H.: Wool keratin-associated protein genes in sheep – a review, Genes, 7, 24, https://doi.org/10.3390/genes7060024, 2016.
Gong, H., Zhou, H., Bai, L., Li, W., Li, S., Wang, J., Luo, Y., and Hickford, J. G. H.: Association of the ovine high glycine-tyrosine keratin-associated protein gene KRTAP20-1 with wool traits, J. Anim. Sci., 97, 587–595, https://doi.org/10.1093/jas/sky465, 2019.
Kijas, J. W., Townley, D., Dalrymple, B. P., Heaton, M. P., Maddox, J. F., McGrath, A., Wilson, P., Ingersoll, R. G., McCulloch, R., and McWilliam, S.: A genome wide survey of SNP variation reveals the genetic structure of sheep breeds, PloS One, 4, e4668, https://doi.org/10.1371/journal.pone.0004668, 2009.
Li, S., Zhou, H., Gong, H., Zhao, F., Wang, J., Liu, X., Luo, Y., and Hickford, J. G. H.: Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits, Genes, 8, E27, https://doi.org/10.3390/genes8010027, 2017a.
Li, S., Zhou, H., Gong, H., Zhao, F., Hu, J., Luo, Y., and Hickford, J. G. H.: Identification of the ovine keratin-associated protein 26-1 (KAP26-1) gene and its association with variation in wool traits, Genes, 8, E225, https://doi.org/10.3390/genes8090225, 2017b.
Li, W., Gong, H., Zhou, H., Wang, J., Liu, X., Li, S., Luo, Y., and Hickford, J. G. H.: Variation in the ovine keratin-associated protein 15-1 gene affects wool yield, J. Agr. Sci., 156, 922–928, https://doi.org/10.1017/S0021859618000953, 2018.
Powell, B. C. and Rogers, G. E.: The role of keratin proteins and their genes in the growth, structure and properties of hair, in: Formation and Structure of Human Hair, edited by: Jollès, P., Zahn, H., and Höcker, H., Birkhäuser Verlag, Basel, Switzerland, 59–148, 1997.
Rogers, G. R., Hickford, J. G. H., and Bickerstaffe, R.: Polymorphism in two genes for B2 high sulfur proteins of wool, Anim. Genet., 25, 407–415, https://doi.org/10.1111/j.1365-2052.1994.tb00531.x, 1994.
Rogers, M. A., Langbein, L., Praetzel-Wunder, S., Winter, H., and Schweizer, J.: Human hair keratin-associated proteins (KAPs), Int. Rev. Cytol., 251, 209–263, https://doi.org/10.1016/S0074-7696(06)51006-X, 2006.
Tao, J., Zhou, H., Gong, H., Yang, Z., Ma, Q., Cheng, L., Ding, W., Li, Y., and Hickford, J. G. H.: Variation in the KAP6-1 gene in Chinese Tan sheep and its effect on wool traits, Small Rumin. Res., 154, 129–132, https://doi.org/10.1016/j.smallrumres.2017.08.001, 2017.
Wang, J., Zhou, H., Zhu, J., Hu, J., Liu, X., Li, S., Luo, Y., and Hickford, J. G. H.: Identification of the ovine keratin-associated protein 15-1 gene (KRTAP15-1) and genetic variation in its coding sequence, Small Rumin. Res., 153, 131–136, https://doi.org/10.1016/j.smallrumres.2017.06.007, 2017.
Zhou, H., Hickford, J. G. H., and Fang, Q.: A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification, Anal. Biochem., 354, 159–161, https://doi.org/10.1016/j.ab.2006.03.042, 2006.
Zhou, H., Gong, H., Yan, W., Luo, Y., and Hickford, J. G. H.: Identification and sequence analysis of the keratin-associated protein 24-1 (KAP24-1) gene homologue in sheep, Gene, 511, 61–65, https://doi.org/10.1016/j.gene.2012.08.049, 2012.
Zhou, H., Gong, H., Li, S., Luo, Y., and Hickford, J. G. H.: A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter, J. Anim. Breed. Genet., 132, 301–307, https://doi.org/10.1111/jbg.12138, 2015.
Zhou, H., Gong, H., Wang, J., Dyer, J. M., Luo, Y., and Hickford, J. G. H.: Identification of four new gene members of the KAP6 gene family in sheep, Sci. Rep., 6, 24074, https://doi.org/10.1038/srep24074, 2016.
Zhang, L., Sun, F., Jin, H., Dalrymple, B. P., Cao, Y., Wei, T., Vuocolo, T., Zhang, M., Piao, Q., and Ingham, A. B.: A comparison of transcriptomic patterns measured in the skin of Chinese fine and coarse wool sheep breeds, Sci. Rep., 7, 14301, https://doi.org/10.1038/s41598-017-14772-4, 2017.
Zhou, H., Visnovska, T., Gong, H., Schmeier, S., Hickford, J. G. H., and Ganley, A. R. D.: Contrasting patterns of coding and flanking region evolution in mammalian keratin associated protein-1 genes, Mol. Phylogenet. Evol., 133, 352–361, https://doi.org/10.1016/j.ympev.2018.12.031, 2019.





