the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Exploring insertions and deletions (indels) of MSRB3 gene and their association with growth traits in four Chinese indigenous cattle breeds
Mingli Wu
Shipeng Li
Guoliang Zhang
Yingzhi Fan
Yuan Gao
Yongzhen Huang
Xianyong Lan
Chuzhao Lei
Ruihua Dang
Methionine sulfoxide reductase B3 (MSRB3) is instrumental in ossification and fat deposition, which regulate the growth and development of cattle directly. The purpose of this study was aimed to explore insertions and deletions (indels) in MSRB3 gene and investigate their association with growth traits in four indigenous cattle breeds (Luxi cattle, Qinchuan cattle, Nanyang cattle, and Jiaxian Red cattle). Four indels were identified by sequencing with DNA pool. Association analysis showed that three of them were associated with growth traits (P<0.05). For P1, the DD (deletion and deletion) genotype was significantly associated with body length of Nanyang cattle; for P6, II (insertion and insertion) and/or DD genotypes were significantly associated with enhanced growth traits of Qinchuan cattle; for P7, II genotype was significantly associated with hucklebone width of Luxi cattle. Our results demonstrated that the polymorphisms in bovine MSRB3 gene were significantly associated with growth traits, which could be candidate loci for marker-assisted selection (MAS) in cattle breeding.
- Article
(1883 KB) - Full-text XML
- BibTeX
- EndNote
There are abundant indigenous cattle breeds in China and most of them were draft cattle for thousands of years (Zhang et al., 2018). Chinese indigenous cattle breeds have the characteristics of slow growth, low yield, and low-grade beef (Yue et al., 2019). With the rapid development of livestock husbandry, beef cattle improvement has become a heated and difficult problem (Singh et al., 2011). However, because of the slow progress in traditional breeding, we need to seek a more efficient and accurate way of breeding urgently (Gui et al., 2015). A noteworthy method is molecular breeding technology, which emerged and developed into an indispensable breeding method, and indels is one of the genetic markers used for assisted selection (Zhang et al., 2003; Fan et al., 2007).
The identification of candidate genes for growth traits has attracted more and more attention. MSRB3 (methionine sulfoxide reductase B3) is an essential member of the MSRB gene family, which can reduce the catalysis effect of methionine-R-sulfoxide to methionine specifically as an oxidoreductase (Kwak et al., 2017a). Previous research has shown that MSRB3 gene could influence ear shape and size, otitis media, heat resistance, oxidation resistance, auditory system, reproductive traits, and size of the hippocampus (Zhang et al., 2017; Fan et al., 2007; Pillas et al., 2010; Shen et al., 2015; Wei et al., 2015; Lee et al., 2012). In recent years, based on the analysis of GWASs (genome-wide association studies), gene silencing, and protein precipitation, MSRB3 was regarded as a candidate gene to affect bovine growth traits (Saatchi et al., 2014). At the same time, it inhibited the proliferation of mouse embryonic fibroblasts and human dermal fibroblasts by acting on the p53, p21, and p27 pathways (Lee et al., 2014; Kwak et al., 2017b).
Consequently, the purpose of this research is to obtain mutation information on MSRB3 gene and its association with the body size of Chinese indigenous cattle through the indel screening and association analysis with growth traits.
All experiments performed in this study were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU). Furthermore, the care and use of animals was fully compliant with local animal welfare laws, guidelines, and policies.
2.1 DNA samples and data collection
A total of 673 Chinese indigenous cattle were used in this study, including Luxi cattle (LX, N=113, Shandong Province), Qinchuan cattle (QC, N=286, Shaanxi Province), Nanyang cattle (NY, N=135, Henan Province), and Jiaxian Red cattle (JX, N=139, Henan Province) (Huang et al., 2017, 2018). All individuals selected were between 2 and 5 years old, healthy, and unrelated. Blood samples were collected in anticoagulant blood vessel and stored at −80 ∘C (Zhao et al., 2017a). In order to demonstrate the relationship between polymorphisms and performance traits, more than 6000 growth traits were measured and calculated, including body weight (BW), body height (BH), body length (BL), chest circumference (ChC), chest depth (ChD), chest width (ChW), hucklebone width (HuW), hip width (HW), cannon circumference (CaC), body length index (BLI), chest circumference index (ChCI), chest width index (ChWI), cannon circumference index (CaCI), hucklebone width index (HuWI), and trunk index (TI) (Zhang et al., 2019; Zhao et al., 2018). All data were measured by the same person following the same standard (Zhao et al., 2018).
2.2 DNA isolation and genomic DNA pools construction
Genomic DNA was isolated from leukocytes and the method refers to a whole-blood genomic DNA extraction kit (Aidlab Biotechnologies Co., Ltd., China). DNA quantity and purity (OD260 ∕ OD280) for each sample was assessed by a NanoDrop™ 1000 spectrometer (Thermo Scientific, Waltham, MA, USA). DNA samples were diluted to a standard concentration (50 ng µL−1) and stored at −80 ∘C. To explore the allele variation of the bovine MSRB3 gene, every 25 samples were mixed and used for polymerase chain reaction (PCR) (Zhao et al., 2017b).
2.3 Indel loci detection and genotyping
Based on the whole-genome sequencing of Chinese indigenous cattle in our lab, seven possible indel loci were found in cattle MSRB3 gene and their PCR primers were designed by Primer Premier 5.0 (Premier Biosoft International, USA) (Table 1) (Chen et al., 2018). PCR amplification was performed in a 20 µL volume system: 1 µL DNA (50 ng µL−1), 10 µL 2× PCR mix (Beijing ComWin Biotech Co., Ltd., China), 0.5 µM of each primers, and 8 µL ddH2O. The amplification conditions of PCR were listed as follows: denaturing at 95 ∘C for 5 min followed by 35 cycles of 95 ∘C for 30 s, annealing for 30 s, and extending at 72 ∘C for 30 s followed by final extension at 72 ∘C for 10 min. PCR products were sequenced directly (Shanghai Sangon Biotech Co., Ltd., P. R. China) to identify potential mutation sites (Zhao et al., 2018). For the confirmed indel loci, every sample was amplified by the aforementioned PCR system and genotyped by 10 % PAGE (polyacrylamide gel electrophoresis) (Pu et al., 2014).
2.4 Statistical analyses
The sequences were contrasted and analyzed by Bioedit software (UK). Hardy–Weinberg equilibrium (HWE), homozygosity (Ho), heterozygosity (He), effective allele numbers (Ne), and polymorphism information content (PIC) were calculated and analyzed using the MSRcall website (http://www.msrcall.com/, last access: 25 August 2018), which was based on Nei's method (Koehl et al., 2018; Yeh et al., 1999; Botstein et al., 1980; Nei, 1973). Distribution differences for genotypic and allelic frequencies among different breeds were analyzed using the χ2 test implemented in SPSS (version 18.0; IBM Corp., Armonk, NY, USA). Linkage disequilibrium was performed by the SHEsis online platform (http://analysis.bio-x.cn, last access: 25 August 2018) (Li et al., 2009). SPSS software (version 18.0) (International Business Machines, US) was used to calculate the associations between different genotypes and growth traits; the structure of the model is , where Yijk is the phenotypic observations, μ is the mean of the phenotypic observations, Gi is the effect of genotype of the jth observation, Bj is the effect of breed in the ith observation, and eijk is the residual effect (Liu et al., 2016; Shi et al., 2016).
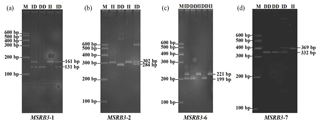
3.1 Variant screening and genotyping of bovine MSRB3 gene
In our previous study, four indel loci (P1, P2, P6, and P7) were identified from seven potential variations in bovine MSRB3 gene. Respectively, P1 was a 30 bp deletion (NC_007303.6 g.48797977-48798006 del GGGAGTAGTTACTGACTGAAGGAAAACATG); P2 was a 18 bp insertion (NC_007303.6 g.48816764-48816765 ins TTCTTTTGGCAACTGCAG); P6 was a 22 bp deletion (NC_007303.6 g.48896074-48896095 del TTTTTCTTTGTCTGGTACACTT); P7 was a 37 bp insertion (NC_007303.6 g.48903091-48903092 ins AGCTGATGTATAACCTCCATAACTTGCTTTCCCCCCT); and no variation was found in P3, P4, or P5 (Fig. 1, 2). All the samples were genotyped by PCR-AFLP (amplified fragment length polymorphism). As shown in the Fig. 1a, wild type (II) showed a 161 bp band, mutant (DD) showed a 131 bp band, and heterozygote (ID) showed 131 and 161 bp bands; as shown in the Fig. 1b, wild type (DD) showed a 284 bp band, mutant (II) showed a 302 bp band, and heterozygote (ID) showed 284 and 302 bp bands; as shown in the Fig. 1c, wild type (II) showed a 221 bp band, mutant (DD) showed a 199 bp band, and heterozygote (ID) showed 199 and 221 bp bands; as shown in the Fig. 1d, wild type (DD) showed a 332 bp band, mutant (II) showed a 369 bp band, and heterozygote (ID) showed 369 and 332 bp bands.
Table 2Genotypes, allele and population genetic information of four indel loci in MSRB3 gene.
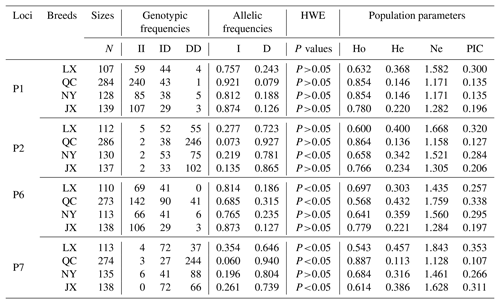
Notes: indel, insertions and deletions; MSRB3, methionine sulfoxide reductase B3; N, number; HWE, Hardy–Weinberg equilibrium; Ho, homozygosity; He, heterozygosity; Ne, effective allele numbers; PIC, polymorphism information content; LX, Luxi cattle; QC, Qinchuan cattle; NY, Nanyang cattle; JX, Jiaxian Red cattle; II, insertion and insertion; ID, insertion and deletion; DD, deletion and deletion.
Table 3The χ2 test of different breeds on four indels of cattle MSRB3 gene.
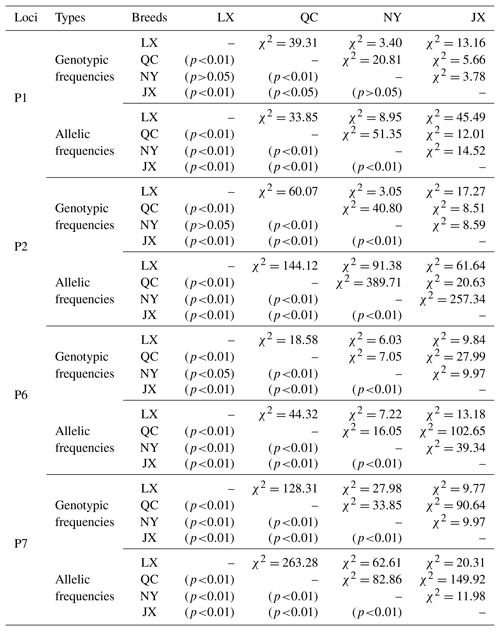
Notes: indel, insertions and deletions; MSRB3, methionine sulfoxide reductase B3; LX, Luxi cattle; QC, Qinchuan cattle; NY, Nanyang cattle; JX, Jiaxian Red cattle.
Table 4Haplotypes and their frequencies of four indels in four Chinese cattle breeds.
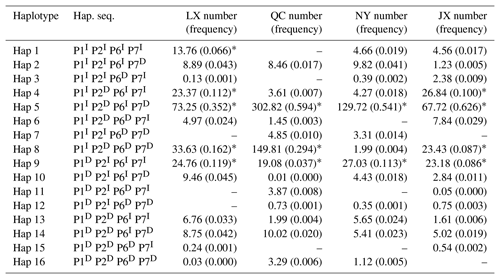
Notes: indel, insertions and deletions; hap, haplotype; hap. seq., haplotype sequence; LX, Luxi cattle; QC, Qinchuan cattle; NY, Nanyang cattle; JX, Jiaxian Red cattle; I, insertion; D, deletion; * means p>0.03.
Table 5Three novel indels in MSRB3 gene and their associations with growth traits.
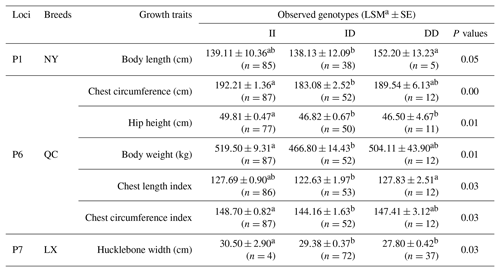
Notes: indel, insertions and deletions; MSRB3, methionine sulfoxide reductase B3; LSM, least squares technique, SE, standard error; LX, Luxi cattle; QC, Qinchuan cattle; NY, Nanyang cattle; II, insertion and insertion; ID, insertion and deletion; DD, deletion and deletion; a, b means P<0.05.
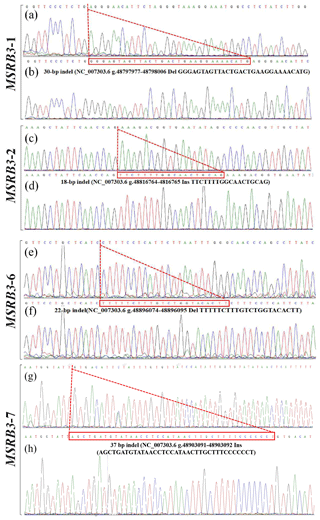
Figure 2Sequencing maps of four indel loci in cattle MSRB3 gene. (a) Homozygotic deletion and deletion type (ID) of MSRB3-1 locus; (b) homozygotic insertion and insertion type (II) of MSRB3-1 locus, the sequence with a dashed red line boundary is a 30 bp deletion; (c) homozygotic deletion and deletion type (ID) of MSRB3-2 locus; (d) homozygotic insertion and insertion type (II) of MSRB3-2 locus, the sequence with a dashed red line boundary is a 18 bp insertion; (e) homozygotic deletion and deletion type (ID) of MSRB3-6 locus; (f) homozygotic insertion and insertion type (II) of MSRB3-6 locus, the sequence with a dashed red line boundary is a 22 bp deletion; (g) homozygotic deletion and deletion type (ID) of MSRB3-7 locus; (h) homozygotic insertion and insertion type (II) of MSRB3-7 locus, the sequence with a dashed red line boundary is a 37 bp insertion.
PCR products showed three genotypes in all of the indel loci in the cattle MSRB3 gene which were detected by 10 % PAGE gel electrophoresis, including (a) the insertion and insertion type (II genotype) consisting of a 161 bp band, deletion and deletion type (DD genotype) consisting of a 131 bp band, and the heterozygote type (ID genotype) that showed 161 and 131 bp bands in MSRB3-1 locus; (b) the insertion and insertion type (II genotype) consisting of a 302 bp band, deletion and deletion type (DD genotype) consisting of a 284 bp band, and the heterozygote type (ID genotype) that showed 302 and 284 bp bands in MSRB3-2 locus; (c) the insertion and insertion type (II genotype) consisting of a 221 bp band, deletion and deletion type (DD genotype) consisting of a 199 bp band, and the heterozygote type (ID genotype) showed 221 and 199 bp bands in MSRB3-6 locus; and (d) the insertion and insertion type (II genotype) consisting of a 369 bp band, deletion and deletion types (DD genotype) consisting of a 332 bp band, and the heterozygote type (ID genotype) that showed 369 and 332 bp bands in MSRB3-7 locus.
3.2 Population genetic variability analysis in four cattle breeds
Genetic diversity is the prerequisite for species survival, adaptation, and evolution, and is also important for improving the performance of meat production. Genotype frequency, allele frequency, and genetic parameters were estimated and shown in Table 2. For P1 locus, the breeds of QC, NY, and JX belonged to a low polymorphic locus (0<PIC<0.25) and LX belonged to a moderate polymorphic locus (0.25<PIC<0.50). All of them were in Hardy–Weinberg equilibrium (HWE) (P>0.05). Homozygosity (Ho) ranged from 0.632 to 0.854. Heterozygosity (He) ranged from 0.146 to 0.368 and effective allele numbers (Ne) were between 1.171 and 1.582. The I allele (0.757–0.921) was higher than D allele (0.079–0.243) in all four breeds. For P2 locus, LX and NY were intermediately polymorphic because they possessed moderate genetic diversity (0.25<PIC<0.50) and QC and JX belonged to low polymorphic locus (0<PIC<0.25). All of them were in Hardy–Weinberg equilibrium (HWE) except NY (P>0.05). Homozygosity (Ho) ranged from 0.600 to 0.864. Heterozygosity (He) ranged from 0.136 to 0.400 and effective allele numbers (Ne) were between 1.158 and 1.668. The I allele (0.073–0.277) was lower than D allele (0.723–0.927) in all four breeds. For P6 locus, LX and QC population deviated from the Hardy-Weinberg equilibrium among the four populations. The populations of LX, QC, and JX belonged to moderate genetic diversity (0.25<PIC<0.50) but the JX population belonged to low polymorphic diversity (0<PIC<0.25). Homozygosity (Ho) ranged from 0.568 to 0.779. Heterozygosity (He) ranged from 0.221 to 0.432 and effective allele numbers (Ne) were between 1.284 and 1.759. The I allele (0.685–0.873) was higher than D allele (0.127–0.315) in all four breeds. For P7, LX, QC, and JX, populations deviated from the Hardy-Weinberg equilibrium, except the NY population. The populations of LX, NY, and JX belonged to moderate genetic diversity (0.25<PIC<0.50) but the QC population belonged to low polymorphic diversity (0<PIC<0.25). Homozygosity (Ho) ranged from 0.543 to 0.887. Heterozygosity (He) ranged from 0.113 to 0.457 and effective allele numbers (Ne) were between 1.128 and 1.843. The I allele (0.060–0.354) was lower than D allele (0.646–0.940) in all four breeds. The χ2 test showed that there was no significant difference in the genotypic frequencies between LX and NY (P1 locus), NY and JX (P1 locus), or LX and NY (P2, P6 loci) (Table 3). In P1 locus, between QC with JX, and in P6 locus, NY with LX populations, the genotypic frequencies were significantly different, and others were extremely and significantly different. Allele frequencies were significantly different among different breeds in four loci (Table 3).
3.3 Haplotype analysis and linkage disequilibrium
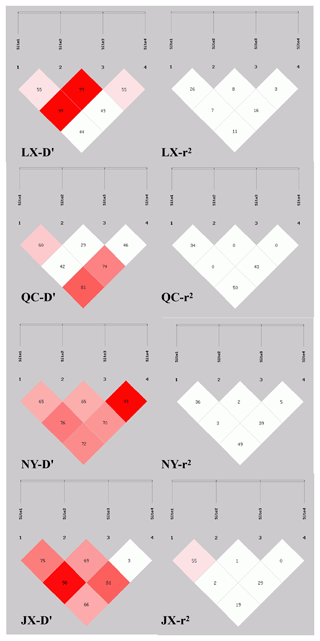
Sixteen haplotypes were analyzed in all four breeds. The frequencies are listed in Table 4, only the frequency of “P1I P2D P6I P7D” and “P1I P2D P6D P7D” haplotypes were all 3 % higher than others in four cattle breeds. The results of linkage disequilibrium of four loci in all four breeds are shown in Fig. 3. Between the P1 and P2, P1 and P4, and P2 and P4 pairs, there is a strong linkage in both QC and NY. In the JX population, between P1 and P2, there is also a strong linkage.
3.4 Association analysis between indels in MSRB3 gene and growth traits
Association analysis, shown in Table 5, revealed that loci of P1, P6, and P7 had a significant effect on growth traits. The chest circumference of DD genotype at P1 locus was significantly longer than ID genotype in the NY population (P<0.05). As for P6 locus, significant differences were found in chest circumference, hip height, and body weight in QC cattle (P<0.01); the II genotype or DD genotype of body length index and chest circumference index showed better performance in the QC population (p<0.05). As for P6 locus, II genotype benefited hucklebone width of the LX population. The linkage disequilibrium showed that P1 and P7 were demonstrated a strong linkage in QC cattle and NY cattle (r2>0.33) (Fig. 3).
In previous studies, the function of MSRB3 is related to ear morphology, auditory system, cell proliferation, and apoptosis (Kwak et al., 2017b; Lee et al., 2014). The results of GWASs have shown that MSRB3 is involved in regulation of ossification and adipose tissue development in cattle (Saatchi et al., 2014). Some studies have shown that the knockout of MSRB3 leads to a decrease in fibroblast proliferation, which could influence the function of proteins, glycosaminoglycan and glycoproteins (Lee et al., 2014). Further study revealed that inhibition of MSRB3 could activate the expression of P53, and enhance the expression level of P21 and P27 subsequently (Kwak et al., 2017b). Above-mentioned studies strongly suggest that MSRB3 inhibited cell proliferation by acting on p53, p21, and p27 pathways, which suggests that MSRB3 is an important candidate gene for animal growth and development (Kwak et al., 2017b; Lee et al., 2014; Eujin et al., 2014). However, there was no study on marker-assisted selection in bovine MSRB3 gene. Therefore, we further verified the relationship between four indel loci in bovine MSRB3 gene and their growth traits.
In our study, P6 was published by NCBI (National Center of Biotechnology Information) and the remaining loci were newly discovered. The χ2 test showed that the P1 was in Hardy-Weinberg equilibrium but P2, P6, and P7 were not in different cattle breeds. The reason may be that artificial selection was carried out to obtain better classification characters, the genetic background of different varieties was inconsistent, or smaller sample size (Doekes et al., 2018; Zhou et al., 2018; Capellini et al., 2017). Through genetic diversity estimation, the classification of polymorphism information content (PIC) resulted in the fact that four loci were in a low polymorphism class in most of the analyzed breeds, the others were identified as having moderate polymorphism. This means that most local cattle have not been bred at these sites. On the basis of two popular measures (r2 and D′) performed in the linkage disequilibrium analysis, all pairs of the P1, P2, and P7 in QC and NY populations demonstrated a strong linkage. Also, a strong linkage between P1 and P2 in the JX population was shown, and others showed a weak linkage. Association analysis shows that three were related to growth traits and there is no evidence that P2 is related to the growth of cattle. P1 caused a 30 bp deletion so as to affect body length in LX. Body length is an important index of meat production; therefore, it can be proved that P1 can be used as a novel marker for breeding (Lacerda et al., 2018; Xu et al., 2018). The deletion of P6 led to a decrease in multiple growth traits of the QC population. Growth traits of II genotype were better than ID and DD genotypes. Both P1 and P6 had harmful mutations. Association analysis of P7 resulted in the fact that individual cattle with II genotype had superior growth traits compared to individuals with ID and DD genotypes. The indels of MSRB3 included functional and nonfunctional types. Functional indels affect the gene product and growth traits, whereas nonfunctional indels may be strongly linked with functional indels of MSRB3.
In a nutshell, the indels of P1, P6, and P7 within MSRB3 gene significantly affected growth traits, meaning that P1, P6, and P7 could be used as DNA markers for eliminating or selecting excellent individuals in marker-assisted selection (MAS) breeding in relation to growth traits, and MSRB3 gene could be used as a candidate gene for breeding beef cattle.
This research identified four indels within MSRB3 gene firstly, which were associated with notable growth traits in four Chinese indigenous cattle breeds. Meanwhile, functional indels could be utilized as promising molecular markers in early marker-assisted selection for cattle breeding after the further extensive research.
Data are available upon request.
RD and MW designed the experiments; SL and GZ collected DNA samples; MW, YF, and YG performed the experiments; YH and XL collected growth data; YM and CL analyzed the experimental data; and RD and MW wrote the paper.
The authors declare that they have no conflict of interest.
This work was supported by the Key Research & Development Plan of Shaanxi Province of China (General Project) (2017NY-071), the Science and Technology Plan of Yangling Demonstration Zone (2018NY-33) and the National Beef Cattle and Yak Industrial Technology System (CARS-37).
This research has been supported by the Key Research & Development Plan of Shaanxi Province of China (grant no. 2017NY-071), the Science and Technology Plan of Yangling Demonstration Zone (grant no. 2018NY-33), and the National Beef Cattle and Yak Industrial Technology System (grant no. CARS-37).
This paper was edited by Steffen Maak and reviewed by Xiaomei Sun and one anonymous referee.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, 1980.
Capellini, T. D., Chen, H., Cao, J., Doxey, A. C., Kiapour, A. M., Schoor, M., and Kingsley, D. M.: Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk, Nat. Genet., 49, 1202–1210, 2017.
Chen, N., Cai, Y., Chen, Q., Li, R., Wang, K., Huang, Y., Hu, S., Huang, S., Zhang, H., Zheng, Z., Song, W., Ma, Z., Ma, Y., Dang, R., Zhang, Z., Xu, L., Jia, Y., Liu, S., Yue, X., Deng, W., Zhang, X., Sun, Z., Lan, X., Han, J., Chen, H., Bradley, D. G., Jiang, Y., and Lei, C.: Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia, Nat. Commun., 9, 2337, https://doi.org/10.1038/s41467-018-04737-0, 2018.
Doekes, H. P., Veerkamp, R. F., Bijma, P., Hiemstra, S. J., and Windig, J. J.: Trends in genome-wide and region-specific genetic diversity in the Dutch-Flemish Holstein-Friesian breeding program from 1986 to 2015, Genet. Sel. Evol., 50, 15, https://doi.org/10.1186/s12711-018-0385-y, 2018.
Eujin, L., GeunHee, K., Kranti, K., and HwaYoung, K.: Methionine sulfoxide reductase B3 deficiency inhibits cell growth through the activation of p53-p21 and p27 pathways, Arch. Biochem. Biophys., 547, 1–5, 2014.
Fan, Y., Wang, W., Ma, G., Liang, L., Shi, Q., and Tao, S.: Patterns of insertion and deletion in Mammalian genomes, Curr. Genomics, 8, 370–378, 2007.
Gui, L., Jiang, B., Zhang, Y., and Zan, L.: Sequence variants in the bovine silent information regulator 6, their linkage and their associations with body measurements and carcass quality traits in Qinchuan cattle, Gene, 559, 16–21, 2015.
Huang, J., Zheng, Q., Wang, S., Zhang, Q., Jiang, L., Hao, R., Li, F., and Ma, Y.: A combined genotype of three SNPs in the bovine gene is related to growth performance in Chinese cattle, Arch. Anim. Breed., 60, 357–362, 2017.
Huang, J., Chen, N., Li, X., An, S., Zhao, M., Sun, T., Hao, R., and Ma, Y.: Two Novel SNPs of PPARγ Significantly Affect Weaning Growth Traits of Nanyang Cattle, Anim. Biotechnol, 29, 68–74, 2018.
Koehl, A. J. and Long, J. C.: The contributions of admixture and genetic drift to diversity among post-contact populations in the Americas, Am. J. Phys. Anthropol., 165, 256–268, https://doi.org/10.1002/ajpa.23347, 2018.
Kwak, G. H. and Kim, H. Y.: MsrB3 deficiency induces cancer cell apoptosis through p53-independent and ER stress-dependent pathways, Arch. Biochem. Biophys., 621, 1–5, 2017b.
Kwak, G. H., Kim, T. H., and Kim, H. Y.: Down-regulation of MsrB3 induces cancer cell apoptosis through reactive oxygen species production and intrinsic mitochondrial pathway activation, Biochem. Biophys. Res. Commun., 483, 468–474, 2017a.
Lacerda, V. V., Campos, G. S., Roso, V. M., Souza, F. R. P., Brauner, C. C., and Boligon, A. A.: Effect of mature size and body condition of Nelore females on the reproductive performance, Theriogenology, 118, 27–33, 2018.
Lee, E., Kwak, G. H., Kamble, K., and Kim, H. Y.: Methionine sulfoxide reductase B3 deficiency inhibits cell growth through the activation of p53-p21 and p27 pathways, Arch. Biochem. Biophys., 547, 1–5, 2014.
Lee, H., Jaffe, A. E., Feinberg, J., Tryggvadottir, R., Brown, S., Montano, C., Aryee, M. J., Irizarry, R. A., Herbstman, J., Witter, F. R., Goldman, L. R., Feinberg, A. P., and Fallin, M. D.: DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth, Int. J. Epidemiol, 41, 188–199, 2012.
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., He, L., and Shi, Y.: A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn), Cell Res., 19, 519–523, 2009.
Liu, M., Li, M., Wang, S., Xu, Y., Lan, X., Li, Z., Lei, C., Yang, D., Jia, Y., and Chen, H.: Association analysis of bovine Foxa2 gene single sequence variant and haplotype combinations with growth traits in Chinese cattle, Gene, 536, 385–392, 2016.
Nei, M.: Analysis of gene diversity in subdivided populations, P. Natl. Acad. Sci. USA, 70, 3321–3323, 1973.
Pillas, D., Hoggart, C. J., Evans, D. M., O'Reilly, P. F., Sipilä, K., Lähdesmäki, R., Millwood, I. Y., Kaakinen, M., Netuveli, G., Blane, D., Charoen, P., Sovio, U., Pouta, A., Freimer, N., Hartikainen, A. L., Laitinen, J, Vaara, S., Glaser, B., Crawford, P., Timpson, N. J., Ring, S. M., Deng, G., Zhang, W., McCarthy, M. I., Deloukas, P., Peltonen, L., Elliott, P., Coin, L. J., Smith, G. D., and Jarvelin, M. R.: Genome-wide association study reveals multiple loci associated with primary tooth development during infancy, PLoS. Genet., 6, e1000856, https://doi.org/10.1371/journal.pgen.1000856, 2010.
Pu, Y., Zhang, Z., Zhou, B., Chen, P., Zhang, K., Song, Y., Gao, Q., Wang, K., Quan, Y., Xi, M., and Zhang, L.: Association of an insertion/deletion polymorphism in IL1A 3'-UTR with risk for cervical carcinoma in Chinese Han Women, Hum. Immunol., 75, 740–744, 2014.
Saatchi, M., Schnabel, R. D., Taylor, J. F., and Garrick, D. J.: Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds, BMC Genomics, 15, 442, https://doi.org/10.1186/1471-2164-15-442, 2014.
Shen, X., Liu, F., Wang, Y., Wang, H., Ma, J., Xia, W., Zhang, J., Jiang, N., Sun, S., Wang, X., and Ma, D.: Down-regulation of msrb3 and destruction of normal auditory system development through hair cell apoptosis in zebrafish, Int. J. Dev. Biol., 59, 195–203, 2015.
Shi, T., Peng, W., Yan, J., Cai, H., Lan, X., Lei, C., Bai, Y., and Chen, H.: A novel 17 bp indel in the SMAD3 gene alters transcription level, contributing to phenotypic traits in Chinese cattle, Arch. Anim. Breed., 59, 151–-157, 2016.
Singh, S., Sharma, S., Arora, J. S., and Sarkhel, B. C.: Molecular and biochemical evaluation of Indian draft breeds of cattle (Bos indicus), Biochem. Genet., 49, 242–250, 2011.
Wei, C., Wang, H., Liu, G., Wu, M., Cao, J., Liu, Z., Liu, R., Zhao, F., Zhang, L., Lu, J., Liu, C., and Du, L.: Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds, BMC Genomics, 16, 194, https://doi.org/10.1186/s12864-015-1384-9, 2015.
Xu, W., He, H., Zheng, L., Xu, J. W., Lei, C. Z., Zhang, G. M., Dang, R. H., Niu, H., Qi, X. L., Chen, H., and Huang, Y. Z.: Detection of 19 bp deletion within PLAG1 gene and its effect on growth traits in cattle, Gene, 675, 144–149, 2018.
Yeh, F. C., Yang, R., and Boyle, T.: POPGENE Version 3.1, Microsoft Window-based Free-ware for Population Genetic Analysis, University of Alberta, Edmonton, AB, Canada 1999.
Yue, B., Wu, J., Shao, S., Zhang, C., Fang, X., Bai, Y., Qi, X., and Chen, H.: Polymorphism in PLIN2 gene and its association with growth traits in Chinese native cattle, Anim. Biotechnol., 5, 1–6, https://doi.org/10.1080/10495398.2018.1555166, 2019.
Zhang, Q., Jin, Y., Jiang, F., Cheng, H., Wang, Y., Lan, X., and Song, E.: Relationship between an indel mutation within the SIRT4 gene and growth traits in Chinese cattle, Anim. Biotechnol., 28, 1-6, https://doi.org/10.1080/10495398.2018.1520716, 2019.
Zhang, W., Gao, X., Zhang, Y., Zhao, Y., Zhang, J., Jia, Y., Zhu, B., Xu, L., Zhang, L., Gao, H., Li, J., and Chen, Y.: Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle, BMC Genet., 19, 114, https://doi.org/10.1186/s12863-018-0705-9, 2018.
Zhang, Z. and Gerstein, M.: Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes, Nucleic Acids Res., 31, 5338–5348, 2003.
Zhang, L. C., Liang, J., Pu, L., Zhang, Y. B., Wang, L. G., Liu, X., Yan, H., and Wang, L. X.: mRNA and protein expression levels of four candidate genes for ear size in Erhualian and Large White pigs, Genet. Mol. Res., 16, 28407177, https://doi.org/10.4238/gmr16029252, 2017.
Zhao, H., He, S., Wang, S., Zhu, Y., Xu, H., Luo, R., Lan, X., Cai, Y., and Sun, X.: Two New Insertion/Deletion Variants of the PITX2 Gene and their Effects on Growth Traits in Sheep, Anim. Biotechnol, 29, 1–7, https://doi.org/10.1080/10495398.2017.1379415, 2017b.
Zhao, H. D., He, S., Zhu, Y. J., Cao, X., Luo, R. Y., Cai, Y., Xu, H. W., and Sun, X. Z.: A novel 29 bp insertion/deletion (indel) variant of the LHX3 gene and its influence on growth traits in four sheep breeds of various fecundity, Arch. Anim. Breed, 60, 79–85, 2017a.
Zhao, H. D., Wu, M. L., Wang, S. H., Yu, X. H., Li, Z., Dang, R. H., and Sun, X. Z.: Identification of a novel 24 bp insertion/deletion (indel) of the androgen receptor gene and its association with growth traits in four indigenous cattle breeds, Arch. Anim. Breed, 61, 71–78, 2018.
Zhou, Z., Li, M., Cheng, H., Fan, W., Yuan, Z., Gao, Q., Xu, Y., Guo, Z., Zhang, Y., Hu, J., Liu, H., Liu, D., Chen, W., Zheng, Z., Jiang, Y., Wen, Z., Liu, Y., Chen, H., Xie, M., Zhang, Q., Huang, W., Wang, W., Hou, S., and Jiang, Y.: An intercross population study reveals genes associated with body size and plumage color in ducks, Nat. Commun., 9, 2648, https://doi.org/10.1038/s41467-018-04868-4, 2018.