the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
DNA methylation of the PLIN1 promoter downregulates expression in chicken lines
Yuhang Sun
Rui Li
Guiying Zhai
Xinyang Zhang
Yuxiang Wang
Evidence suggests that Perilipin-1 (PLIN1) is subject to functional regulation by epigenetic modifications in women with obesity. However, whether chicken PLIN1 expression is regulated by DNA methylation is unknown. Here, Sequenom MassARRAY and real-time polymerase chain reaction (PCR) were conducted to analyze the promoter methylation status and expression of the PLIN1 gene in Northeast Agricultural University broiler lines divergently selected for abdominal fat content. We found that chicken PLIN1 expression was significantly higher in adipose tissue of fat-line broilers than in lean lines at 1–7 weeks of age, and was significantly positively correlated with abdominal fat percentage (AFP) in chicken adipose development (Pearson's r=0.627, P<0.001). The region analyzed for DNA methylation was from −12 to −520 bp upstream of the translation start codon ATG, and had five CpG sites, where only the DNA methylation levels of CpG5 located at position −490 bp were significantly higher in lean compared to fat chickens at 5 and 6 weeks (P<0.05) and were significantly negatively correlated with PLIN1 mRNA levels and AFP (P<0.05). These results shed new light on the regulation of hypertrophic growth in chicken adipose development.
- Article
(907 KB) - Full-text XML
-
Supplement
(109 KB) - BibTeX
- EndNote
Obesity is characterized by an expansion of white adipose tissue (WAT) mass resulting from increased adipocyte number and/or size. One of the most important components of mature adipocytes is lipid droplets with intracellular space almost occupied by lipid droplets. The degree of adipocyte differentiation mainly depends on the size of lipid droplets. Perilipin-1 (PLIN1), a lipid-droplet-associated protein, was originally identified in adipocytes (Greenberg et al., 1991). Our group showed that lipid droplets are surrounded by PLIN1 in chicken adipocytes at different time points after cell differentiation (Qin et al., 2016). In the basic condition, overexpression of PLIN1 promotes chicken preadipocyte lipid accumulation (Zhou et al., 2012). With hormone stimulation, overexpression of PLIN1 inhibits lipid accumulation in adipocyte cells, consistent with findings in mammals (Miyoshi et al., 2008, 2007, 2006).
Obesity is associated with increased basal lipolysis and decreased levels of PLIN1 protein in adipose tissue (Mottagui-Tabar et al., 2003; Ray et al., 2009; Wang et al., 2003). Evidence suggests that PLIN1 is subject to functional regulation by epigenetic modifications in women with obesity (Bialesova et al., 2017). However, the DNA methylation status of the PLIN1 gene and its role in chicken adipose development has not been elucidated. To enhance our understanding of molecular mechanisms underlying chicken adipose tissue development and adipogenesis, investigating the DNA methylation status of PLIN1 and its effect on chicken adipose development is essential.
Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) have been selected by long-term divergent selection since 1996 using abdominal fat percentage (AFP) and plasma very low-density lipoprotein (VLDL) concentration (Liu et al., 2007). After 19 generations of selection, differences in AFP and abdominal fat weight (AFW) were striking. PLIN1 is a critical regulator of fat storage and breakdown and its expression is significantly higher in adipose tissue of fat-line broilers than lean broilers at 7 weeks (Wang et al., 2011). This result suggests a significant association between PLIN1 expression and abdominal fat content. DNA methylation is important for regulation of gene expression and is critical in establishing patterns of gene repression during development (Cedar and Bergman, 2009). We hypothesized that the chicken PLIN1 gene is differently methylated in the lean and fat chicken lines during adipose tissue development.
Here, we used Sequenom MassARRAY and real-time PCR to analyze the promoter methylation status and expression of the PLIN1 gene in abdominal adipose of lean and fat chicken lines at 1–7 weeks of age. Our findings showed a positive correlation between AFP and PLIN1 mRNA levels in chicken adipose development, and DNA methylation levels of CpG5 were significantly higher in lean compared to fat chickens at 5 and 6 weeks and were significantly negatively correlated with PLIN1 mRNA levels and AFP. This result suggests that epigenetic regulation of PLIN1 might be important for hypertrophic growth in chicken adipose development.
2.1 Animals and tissues
Animal work was carried out in accordance with the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People's Republic of China (approval number: 2006-398) and the Laboratory Animal Management Committee of Northeast Agricultural University (Harbin, P. R. China). Chickens from NEAUHLF generation 19 (G19) were used. All birds were housed under similar environmental conditions with free access to feed and water. The abdominal fat tissues, abdominal fat pad and adipose tissue surrounding the gizzard from each individual male bird were stripped and weighed as AFW, then snap-frozen in liquid nitrogen and stored at −80∘ for extraction of genomic DNA and total RNA. A total of 70 male birds (five birds per line per time point) at 1–7 weeks of age were used in this process. AFP = AFW/body weight.
2.2 RNA isolation and quantitative real-time RT-PCR
Total RNA from abdominal adipose tissues was extracted by using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol and treated with DNase I (Takara, Dalian, China), via visualization of the 18S and 28S ribosomal RNA bands on a denaturing formaldehyde agarose gel to assess RNA quality. Complementary DNAs were synthesized in a final volume of 20 µL with 1 mg of total RNA, an oligo(dT) anchor primer and ImProm-II reverse transcriptase (Promega, Madison, WI, USA). Conditions were 25 ∘C for 5 min, 42 ∘C for 60 min and 70 ∘C for 15 min.
Quantitative relative-transcription PCR (RT-PCR) was performed with the 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using FastStart Universal SYBR Green Master kits (Roche, Shanghai, China). From each RT reaction, within a 10 µL reaction add 1 µL of product; reaction mixtures were incubated in an ABI Prism 7500 sequence detection system (Applied Biosystems) at one cycle at 95 ∘C for 10 min, 40 cycles of 95 ∘C for 15 s and 60 ∘C for 1 min. To detect and eliminate possible primer-dimer artifacts, we using Dissociation Curve 1.0 software (Applied Biosystems) to analyze the dissociation curves for each PCR reaction. The relative expression of PLIN1 was calculated using the formula 2Δ−CT with TATA-box binding protein (TBP) as an internal reference. Primers used for quantitative RT-PCR were shown in Table 1.
2.3 Methylation analysis
The Sequenom MassARRAY platform was used to determine methylation levels of loci in the CpG island promoter of selected PLIN1 genes (GeneBank accession no. GU327532.1). CpG islands were predicted using CpG Island Searcher software (https://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/, last access: 3 June 2019). Thresholds were GC > 50 %, CpG observed ∕ expected value >0.6 and CpG island length >200 bp.
Adipose tissue DNA was extracted and isolated with QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. NanoDrop spectrophotometer from GE Healthcare Life Science (Uppsala, Sweden) was conducted to quantify the genomic DNA. Bisulfite conversation of the DNA was performed using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. The online software EpiDesigner (https://www.epidesigner.com,last access: 3 June 2019, Agena Bioscience, USA) was used to design PCR primers, and primer sequences were shown in Table 1. PCR products were used as a template for transcription and base-specific cleavage reactions using the MassCLEAVE kit (Sequenom, USA). DNA methylation levels of fragmented samples were quantified by a MassARRAY analyzer compact matrix-assisted laser desorption/ionization time-of-flight mass spectrometry instrument and EpiTYPER analyzer software (Sequenom). Individual CpG sites or clusters of consecutive CpG sites were defined as CpG units following the manufacturer's protocol.
2.4 Statistical analysis
All data are presented as means plus standard error (SE). Differences between groups were analyzed using unpaired, two-tailed Student's t tests. Pearson's r was used to analyze the correlation between AFP and mRNA levels, methylation and mRNA levels, and AFP and methylation. P<0.05 was considered significant.
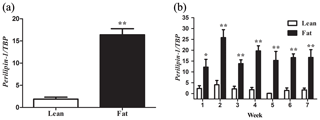
Figure 1PLIN1 expression in NEAUHLF abdominal adipose tissues. The mRNA levels were determined by real-time quantitative reverse-transcription polymerase chain reaction and normalized to TATA-box binding protein (TBP) mRNA measured in parallel experiments. Results are the mean ± SE. (a) Mean PLIN1 expression in adipose tissues of lean and fat broilers (n=35). (b) Mean PLIN1 expression in adipose tissues of lean and fat broilers at 1–7 weeks of age (n=5). and .
Table 2AFW and AFP in lean and fat lines of NEAUHLF.
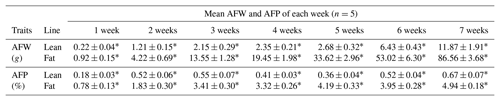
Comparison of lean and fat lines using unpaired Student's t test. Values are the mean ± SE. ∗ Significant difference in AFW and AFP between the two chicken lines (P<0.01).
3.1 PLIN1 and abdominal fat deposition
We used 70 male birds that had normal weight in all test weeks of age (Fig. S1 in the Supplement). AFP and AFW were calculated in G19 (Table 2). AFW and AFP were significantly different between the two lines in adipose development and AFP in the fat line at 7 weeks of age was 7.37 times higher than the lean line (Table 2). In our previous study, in G11, PLIN1 expression was significantly higher in adipose tissue of fat-line broilers than lean broilers at 7 weeks (Wang et al., 2011). To study if PLIN1 expression was related to AFP, the relationship between PLIN1 mRNA levels in abdominal fat tissue and abdominal fat content was analyzed. PLIN1 transcript levels in G19 fat males were higher than in lean males at 1–7 weeks of age (Fig. 1) (P<0.05). With increased age, no significant difference was seen in PLIN1 expression, which maintained a relatively low level in the lean line (Fig. 1b). Correlation analysis showed a significant positive correlation between AFP and PLIN1 mRNA levels in chicken adipose development (Pearson's r=0.627, P<0.001).
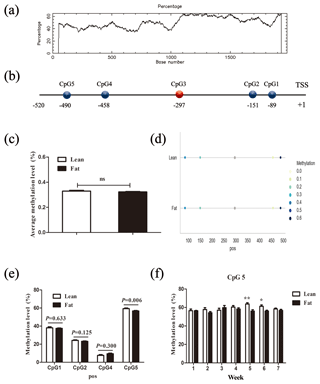
Figure 2Comparison of CpG site methylation at the chicken PLIN1 promoter in NEAUHLF. (a) Search for CpG islands in the chicken PLIN1 promoter. Thresholds were GC >50 %, CpG observed/expected value >0.6 and CpG island length >200 bp. (b) Schematic diagram of PLIN1 gene promoter. All numbered positions are relative to the adenine of the translation start site of chicken PLIN1. Five CpG sites were found in the analyzed region, in which CpG3 (red) was not detected. (c) Average methylation level of promoter region in line and fat lines. Panels (d) and (e) show average methylation levels of the PLIN1 promoter CpG sites in adipose tissues of lean and fat broilers in all weeks (n=35). (f) Average methylation levels of CpG5 in adipose tissues of lean and fat broilers at 1–7 weeks of age (n=5). and .
3.2 DNA methylation of chicken PLIN1 promoter in adipose tissue
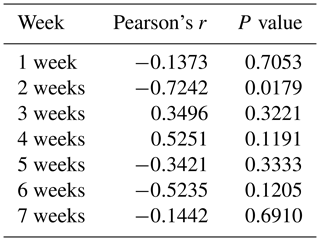
To determine whether the PLIN1 gene expressed in chicken abdominal adipose tissue is regulated by DNA methylation, we used CpG Searcher software. Bioinformatics analysis revealed that the 2 kb promoter region of chicken PLIN1 had no typical CpG islands (Fig. 2a). Then, we investigated the methylation status within a 509 bp region from −520 to −12 bp upstream of the translation start codon ATG that contained PLIN1 core promoter regions (Zhou et al., 2016). Five CpG sites were found in the analyzed region, at positions −89, −151, −297, −458 and −490 bp (Fig. 2b). The sites were named CpG1 through CpG5. We quantified the methylation levels of CpG units within the promoter using Sequenom MassARRAY technology. For technical reasons, the CpG3 site (red, Fig. 2b) could not be detected. In summary, the average methylation level of the promoter region, with five CpG sites, was not significantly different in the lean line compared to the fat line (Fig. 2c). Then, we tested the relationship between the average methylation level of promoter region and PLIN1 expression level at all tested ages, and only the promoter methylation at 2 weeks of age showed a significantly negative correlation with PLIN1 expression (Pearson's , P=0.018) (Table 3).
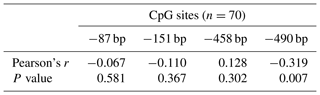
Single CpG site methylation analysis showed that hypermethylated CpG sites were not found in the two lines in the PLIN1 promoter region, and only average DNA methylation levels of CpG5 at position −490 bp were significantly higher in lean than fat chickens (P=0.006) (Fig. 2d, e). Further analysis showed that CpG5-site methylation levels were significantly higher in the lean line than in the fat line at 5 and 6 weeks (P<0.05, Fig. 2f). Then, we tested correlations between average DNA methylation level of each CpG site and PLIN1 expression level. The results showed that, of all tested CpGs, only the average methylation level of CpG5 at −490 bp displayed a significantly negative correlation with PLIN1 expression level (Pearson's , P=0.007) (Table 4), and was negatively correlated with AFP at 5 and 6 weeks of age (Pearson's , P<0.001 and Pearson's , P=0.047, respectively).
Studies suggest that PLIN1 is highly expressed in white adipocytes and is actively involved in lipolysis regulation through interaction with hormone-sensitive lipase and lipase activator CGI-58 (adipose triglyceride lipase) (Contreras et al., 2017; Granneman et al., 2009). PLIN1 knockout increases basal lipolysis and decreases liquid droplet (LD) size in adipocytes and causes resistance to diet-induced obesity in mice (Martinez-Botas et al., 2000; Tansey et al., 2001). PLIN1 mice are lean, with normal body weight but reduced WAT stores. Furthermore, PLIN1 mice are resistant to diet and genetically induced obesity (Castro-Chavez et al., 2003; Greenberg et al., 1991; Martinez-Botas et al., 2000; Saha et al., 2004; Tansey et al., 2001).
Our results indicated that PLIN1 expression was significantly higher in fat lines than in lean lines at all tested ages. Our results also demonstrated a significant positive correlation between AFP and mRNA expression in chicken adipose development. This result indicated that PLIN1 might be a marker gene for selection for fatness.
Adipocyte gene transcription is modulated by epigenetic mechanisms. Chicken PPARγ is regulated by DNA methylation during adipose tissue development (Sun et al., 2014) and DNA methylation may regulate CEBPA expression in early chicken adipose development (Gao et al., 2015). Dysregulated CpG methylation of lipolysis genes is a major feature of the adipocyte epigenetic signature in women with obesity and epigenetic regulation of PLIN1 is important for increased adipocyte lipolysis in insulin-resistant states such as obesity (Bialesova et al., 2017). In this study, we did not find a CpG island in the upstream 2.0 kb of the translation start codon of chicken PLIN1. Only in four CpG loci detected in the core promoter region of the gene, did we find that CpG5 was negatively correlated with chicken PLIN1 mRNA expression, and the DNA methylation level of this locus was negatively correlated with AFP in broilers at 5 and 6 weeks of age. Studies on the development of adipose tissue in chickens show that increases in the abdominal fat pad mass of broiler chickens mainly depends on hyperplasia of adipocytes until 4 weeks of age and hypertrophic growth beyond 4 weeks (Hood, 1982). PLIN1 can augment triglyceride synthesis and promote enlargement of lipid droplets (LDs), leading to the formation of large LDs (Koltes and Spurlock, 2011; Sun et al., 2013). Therefore, we speculate that DNA methylation can negatively regulate the expression of the PLIN1 gene during the growth and development of adipose tissue in broiler chickens, affect the hypertrophy of adipocytes and then inhibit the accumulation of body fat. In addition, we also found that adding methyltransferase inhibitors significantly increased the expression of the PLIN1 gene in DF-1 cells (Date not listed). In short, the influences of promoter DNA methylation on PLIN1 gene expression shed new light on the regulation of hypertrophic growth in chicken adipose development.
Future work is needed to define how CpG methylation interacts with other known regulators of PLIN1 mRNA expression. Mammalian studies show that DNA methylation, including hypermethylation and hypomethylation, are important for regulating the expression of transcription factors, transcriptional cofactors and other genes involved in mammalian adipose development and adipogenesis (Bowers et al., 2006; Noer et al., 2007; Shore et al., 2010). CpG methylation at promoter regions has been widely recognized as an effective epigenetic modification, which prohibits transcription factor recruitment, resulting in transcription suppression (Hu et al., 2013). We used TFBIND (http://tfbind.hgc.jp/, last access: 3 June 2019) and JASPAR (http://jaspar.genereg.net/, last access: 3 June 2019) and to predict transcription factor binding sites around the promoter region detected in this study. TFBIND results showed that the CpG5 site match reported binding motifs for NF-κB and E2F family members. NF-κB is the well-known transcription factor regulating PLIN1 expression (Laurencikiene et al., 2007). Recent evidence suggests that a cytosine at the −1 position of a κB site (−1C) could be methylated, which thereafter impaired NF-κB binding and/or function (Wang et al., 2017). However, it is unknown whether the methylation of CpG5 will affect NF-κB's binding to its binding site. CpG methylation differentially regulates the response of certain E2F elements to different E2F family members (Campanero et al., 2000). The E2F consensus motif contained only one methylation CpG and did not affect binding of E2F2-5, but abrogated E2F1's binding (Campanero et al., 2000). Therefore, the CpG5 site may be an E2F1 binding site and methylation of CpG might influence E2F1's function. However, to date, there is no report of E2F1 regulating PLIN1 expression.
In addition, multiple binding motifs for transcription factors, such as C/EBPβ, NRF1, KLF4 and KLF5, were found around the CpG5 site (±30 bp). Evidence suggests that KLF4 can activate C/EBPβ expression, and C/EBPβ and KLF5 can co-activate PPARγ expression and promote adipocyte differentiation (Oishi et al., 2005; Birsoy et al., 2008). NRF1 has been implicated in lipid droplet accumulation, negative regulation of the P1 promoter of PPARγ gene, and inhibition of chicken adipogenesis (Cui et al., 2018; Liu et al., 2008). Although those transcription factors have effects on adipocyte differentiation, CpG5 prevents or promotes which transcription factors' bind to influence the expression regulating of PLIN1 gene. Further research is needed.
Taken together, in this study, we suggested that the PLIN1 gene was a marker gene for selection of fat traits. The DNA methylation of CpG5 at position −490 bp of the PLIN1 promoter has a certain impact on PLIN1 gene expression. Our results imply that epigenetic regulation of PLIN1 might be important for hypertrophic growth in chicken adipose development.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-62-375-2019-supplement.
The authors declare that they have no conflict of interest.
We thank International Science Editing (http://www.internationalscienceediting.com, last access: 21 November 2018) for editing a previous version of this paper.
This research has been supported by the National Basic Research Program of China (Youth Science Foundation) project (grant no. 31201796), the special research foundation for doctoral science points in colleges and universities (grant no. 20122325120008), and the Academic Backbone Project of Northeast Agricultural University (grant no. 16XG13).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Bialesova, L., Kulyte, A., Petrus, P., Sinha, I., Laurencikiene, J., Zhao, C., Wright, K. D., Arner, P., and Dahlman, I.: Epigenetic regulation of PLIN 1 in obese women and its relation to lipolysis, Sci. Rep., 7, 10152, https://doi.org/10.1038/s41598-017-09232-y, 2017.
Birsoy, K., Chen, Z., and Friedman, J.: Transcriptional regulation of adipogenesis by KLF4, Cell Metab., 7, 339–347, https://doi.org/10.1016/j.cmet.2008.02.001, 2008.
Bowers, R. R., Kim, J. W., Otto, T. C., and Lane, M. D.: Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene, P. Natl. Acad. Sci. USA, 103, 13022–13027, https://doi.org/10.1073/pnas.0605789103, 2006.
Campanero, M. R., Armstrong, M. I., and Flemington, E. K.: CpG methylation as a mechanism for the regulation of E2F activity, P. Natl. Acad. Sci. USA, 97, 6481–6486, https://doi.org/10.1073/pnas.100340697, 2000.
Castro-Chavez, F., Yechoor, V. K., Saha, P. K., Martinez-Botas, J., Wooten, E. C., Sharma, S., O'Connell, P., Taegtmeyer, H., and Chan, L.: Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in perilipin knockout mice: a microarray gene expression profile, Diabetes., 52, 2666–2674, 2003.
Cedar, H. and Bergman, Y.: Linking DNA methylation and histone modification: patterns and paradigms, Nat. Rev. Genet., 10, 295–304, https://doi.org/10.1038/nrg2540, 2009.
Contreras, G. A., Strieder-Barboza, C., and Raphael, W.: Adipose tissue lipolysis and remodeling during the transition period of dairy cows, J. Anim. Sci. Biotechnol., 8, 41, https://doi.org/10.1186/s40104-017-0174-4, 2017.
Cui, T., Xing, T., Huang, J., Mu, F., Jin, Y., You, X., Chu, Y., Li, H., and Wang, N.: Nuclear Respiratory Factor 1 Negatively Regulates the P1 Promoter of the Peroxisome Proliferator-Activated Receptor-gamma Gene and Inhibits Chicken Adipogenesis, Front Physiol., 9, 1823, https://doi.org/10.3389/fphys.2018.01823, 2018.
Dalen, K. T., Schoonjans, K., Ulven, S. M., Weedon-Fekjaer, M. S., Bentzen, T. G., Koutnikova, H., Auwerx, J., and Nebb, H. I.: Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma, Diabetes., 53, 1243–1252, 2004.
Gao, Y., Sun, Y., Duan, K., Shi, H., Wang, S., Li, H., and Wang, N.: CpG site DNA methylation of the CCAAT/enhancer-binding protein, alpha promoter in chicken lines divergently selected for fatness, Anim. Genet., 46, 410–417, https://doi.org/10.1111/age.12326, 2015.
Granneman, J. G., Moore, H. P., Krishnamoorthy, R., and Rathod, M.: Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl), J. Biol. Chem., 284, 34538–34544, https://doi.org/10.1074/jbc.M109.068478, 2009.
Greenberg, A. S., Egan, J. J., Wek, S. A., Garty, N. B., Blanchette-Mackie, E. J., and Londos, C.: Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets, J. Biol. Chem., 266, 11341–11346, 1991.
Hood, R. L.: The cellular basis for growth of the abdominal fat pad in broiler-type chickens, Poult. Sci., 61, 117–121, https://doi.org/10.3382/ps.0610117, 1982.
Hu, S., Wan, J., Su, Y., Song, Q., Zeng, Y., Nguyen, H. N., Shin, J., Cox, E., Rho, H. S., Woodard, C., Xia, S., Liu, S., Lyu, H., Ming, G. L., Wade, H., Song, H., Qian, J., and Zhu, H.: DNA methylation presents distinct binding sites for human transcription factors, Elife., 2, e726, https://doi.org/10.7554/eLife.00726 2013.
Koltes, D. A. and Spurlock, D. M.:. Coordination of lipid droplet-associated proteins during the transition period of Holstein dairy cows, J. Dairy. Sci., 94, 1839–1848, https://doi.org/10.3168/jds.2010-3769, 2011.
Laurencikiene, J., van Harmelen, V., Arvidsson, Nordstrom E., Dicker, A., Blomqvist, L., Naslund, E., Langin, D., Arner, P., and Ryden, M.: NF-kappaB is important for TNF-alpha-induced lipolysis in human adipocytes, J. Lipid. Res., 48, 1069–1077, https://doi.org/10.1194/jlr.M600471-JLR200, 2007.
Liu, S., Wang, S. Z., Li, Z. H., and Li, H.: Association of single nucleotide polymorphism of chicken uncoupling protein gene with muscle and fatness traits, J. Anim. Breed. Genet., 124, 230–235, https://doi.org/10.1111/j.1439-0388.2007.00654.x, 2007.
Liu, Y., Niu, N., Zhu, X., Du, T., Wang, X., Chen, D., Wu, X., Gu, H. F., and Liu, Y.: Genetic variation and association analyses of the nuclear respiratory factor 1 (nRF1) gene in Chinese patients with type 2 diabetes, Diabetes., 57, 777–782, https://doi:10.2337/db07-0008, 2008.
Livak, K. J. and Schmittgen, T. D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods., 25, 402–408, https://doi.org/10.1006/meth.2001.1262, 2001.
Martinez-Botas, J., Anderson, J. B., Tessier, D., Lapillonne, A., Chang, B. H., Quast, M. J., Gorenstein, D., Chen, K. H., and Chan, L.: Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice, Nat. Genet., 26, 474–479, https://doi.org/10.1038/82630, 2000.
Martinez-Botas, J., Anderson, J. B., Tessier, D., Lapillonne, A., Chang, B. H., Quast, M. J., Gorenstein, Miyoshi, H., Perfield, J. W., Obin, M. S., and Greenberg, A. S.: Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes, J. Cell. Biochem., 105, 1430–1436, https://doi.org/10.1002/jcb.21964, 2008.
Miyoshi, H., Souza, S. C., Zhang, H. H., Strissel, K. J., Christoffolete, M. A., Kovsan, J., Rudich, A., Kraemer, F. B., Bianco, A. C., Obin, M. S., and Greenberg, A. S.: Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and independent mechanisms, J. Biol. Chem., 281, 15837–15844, https://doi.org/10.1074/jbc.M601097200, 2006.
Miyoshi, H., Perfield, J. W., Souza, S. C., Shen, W. J., Zhang, H. H., Stancheva, Z. S., Kraemer, F. B., Obin, M. S., and Greenberg, A. S.: Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes, J. Biol. Chem., 282, 996–1002, https://doi.org/10.1074/jbc.M605770200, 2007.
Mottagui-Tabar, S., Ryden, M., Lofgren, P., Faulds, G., Hoffstedt, J., Brookes, A. J., Andersson, I., and Arner, P.: Evidence for an important role of perilipin in the regulation of human adipocyte lipolysis, Diabetologia., 46, 789–797, https://doi.org/10.1007/s00125-003-1112-x, 2003.
Noer, A., Boquest, A. C., and Collas, P.: Dynamics of adipogenic promoter DNA methylation during clonal culture of human adipose stem cells to senescence, BMC. Cell. Biol., 8, 18, https://doi.org/10.1186/1471-2121-8-18, 2007.
Oishi, Y., Manabe, I., Tobe, K., Tsushima, K., Shindo, T., Fujiu, K., Nishimura, G., Maemura, K., Yamauchi, T., Kubota, N., Suzuki, R., Kitamura, T., Akira, S., Kadowaki, T., and Nagai, R.: Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation, Cell Metab., 1, 27–39, https://10.1016/j.cmet.2004.11.005, 2005.
Qin, F. Y., Zhou, W. N., Wang, Y. B., Cheng, B. H., Li, H., and Wang, Y. X.: Cloning and subcellular distribution of chicken (Gallus gallus) Perilipin1 gene, Journal of Agricultural Biotechnology, 24, 1560–1568, https://doi.org/10.3969/j.issn.1674-7968, 2016.
Ray, H., Pinteur, C., Frering, V., Beylot, M., and Large, V.: Depot-specific differences in perilipin and hormone-sensitive lipase expression in lean and obese, Lipids. Health. Dis., 8, 58, https://doi.org/10.1186/1476-511X-8-58, 2009.
Saha, P. K., Kojima, H., Martinez-Botas, J., Sunehag, A. L., and Chan, L.: Metabolic adaptations in the absence of perilipin: increased beta-oxidation and decreased hepatic glucose production associated with peripheral insulin resistance but normal glucose tolerance in perilipin-null mice, J. Biol. Chem., 279, 35150–35158, https://doi.org/10.1074/jbc.M405499200, 2004.
Shore, A., Karamitri, A., Kemp, P., Speakman, J. R., and Lomax, M. A.: Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue, Diabetologia, 53, 1164–1173, https://doi.org/10.1007/s00125-010-1701-4, 2010.
Stenson, B. M., Ryden, M., Venteclef, N., Dahlman, I., Pettersson, A. M., Mairal, A., Astrom, G., Blomqvist, L., Wang, V., Jocken, J. W., Clement, K., Langin, D., Arner, P., and Laurencikiene, J.: Liver X receptor (LXR) regulates human adipocyte lipolysis, J. Biol. Chem., 286, 370–379, https://doi.org/10.1074/jbc.M110.179499, 2011.
Sun, Z., Gong, J., Wu, H., Xu, W., Wu, L., Xu, D., Gao, J., Wu, J. W., Yang, H., Yang, M., and Li, P.: Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes, Nat. Commun., 4, 1594, https://doi.org/10.1038/ncomms2581, 2013.
Sun, Y. N., Gao, Y., Qiao, S. P., Wang, S. Z., Duan, K., Wang, Y. X., Li, H., and Wang, N.: Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor gamma in chicken lines divergently selected for fatness, J. Anim. Sci., 92, 48–53, https://doi.org/10.2527/jas.2013-6962, 2014.
Tansey, J. T., Sztalryd, C., Gruia-Gray, J., Roush, D. L., Zee, J. V., Gavrilova, O., Reitman, M. L., Deng, C. X., Li, C., Kimmel, A. R., and Londos, C.: Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity, P. Natl. Acad. Sci. USA, 98, 6494–6499, https://doi.org/10.1073/pnas.101042998, 2001.
Wang, T., Li, J., Ding, K., Zhang, L., Che, Q., Sun, X., Dai, Y., Sun, W., Bao, M., Wang, X., Yang, L., and Li, Z.: The CpG Dinucleotide Adjacent to a kappaB Site Affects NF-kappaB Function through Its Methylation, Int. J. Mol. Sci., 18, 528, https://doi.org/10.3390/ijms18030528, 2017.
Wang, Y., Sullivan, S., Trujillo, M., Lee, M. J., Schneider, S. H., Brolin, R. E., Kang, Y. H., Werber, Y., Greenberg, A. S., and Fried, S. K.: Perilipin expression in human adipose tissues: effects of severe obesity, gender, and depot, Obes. Res., 11, 930–936, https://doi.org/10.1038/oby.2003.128, 2003.
Wang, Y. B., Wang, N., Wang, L., Wang, Y. X., Li, Y. M., and Li, H.: Preparation of antiserums against chicken Perilipin1 and tissue expression analyses of Perilipin1, Chinese Journal of Animal and Veterinary Sciences, 42, 349–355, 2011.
Zhou, W. N., Wang, Y. X., and Li, H.: Effect of perilipin 1 on chicken preadipocyte lipid accumulation, Chinese Journal of Cellular and Molecular Immunology, 28, 944–947, https://doi.org/10.13423/j.cnki.cjcmi.006537, 2012.
Zhou, W. N., Shi, M. X., Qiao, S. P., Shi, H. Y., and Wang, Y. X.: Promoter cloning and analysis of chicken Perilipin1 gene, Chinese Journal of Animal and Veterinary Sciences, 47, 249–259, https://doi.org/10.11843/j.issn.0366-6964, 2016.