the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Genetic diversity of different breeds of Kazakh sheep using microsatellite analysis
Kairat Dossybayev
Zarina Orazymbetova
Aizhan Mussayeva
Naruya Saitou
Rakhymbek Zhapbasov
Bolathan Makhatov
Bakytzhan Bekmanov
A total of 75 individuals from five sheep populations in Kazakhstan were investigated based on 12 STR (short tandem repeat, also known as microsatellite) markers in order to study their genetic structure and phylogenetic relationship based on genetic distances. These sheep had a high level of genetic diversity. In total, 163 alleles were found in all the populations using 12 microsatellite loci. The mean number of alleles, effective number of alleles, and polymorphism information content (PIC) values per loci were 13.4, 5.9, and 0.78, respectively. Comparing the allelic diversity between the populations, the highest genetic diversity was observed in the Edilbay-1 sheep breed (8.333±0.644), and the lowest parameter was for Kazakh Arkhar-Merino (7.083±0.633). In all populations, there is a deficiency of heterozygosity. The largest genetic diversity was found in loci INRA023 and CSRD247 with 16 alleles, and the smallest polymorphism was noted for the locus D5S2 with 8 alleles. The level of observed heterozygosity was in the range 0.678±0.051 for Kazakh Arkhar-Merino and 0.767±0.047 for Kazakh fat-tailed coarse wool. The expected heterozygosity level range was from 0.702±0.033 for Kazakh Arkhar-Merino to 0.777±0.023 for Edilbay-1. When 12 microsatellite loci are compared, the OarFCB20 locus showed the highest level of genetic variability. Excess of heterozygosity was observed at three loci; MAF065, McM042, and OarFCB20. The highest genetic distance was observed between Kazakh Arkhar-Merino and Edilbay-1, whereas the genetic distance between Edilbay-1 and Edilbay-2 is the smallest using Nei's standard genetic distance. The Edilbay-1 sheep breed possesses the largest genetic diversity among these five populations.
- Article
(406 KB) - Full-text XML
- BibTeX
- EndNote
Sheep breeding is the most ancient branch of animal husbandry in Kazakhstan. The country has more than 20 distinct sheep breeds to date. Among them, the sheep breeds which first appeared in Kazakhstan by origin and history are Edilbay, Kazakh Arkhar-Merino, Kazakh Finewool, and Kazakh fat-tailed coarse wool. These breeds are well adapted to the various climatic conditions in Kazakhstan. Various breeds of sheep in one way or another are different in terms of biological efficiency. This study aims at investigating the genetic diversity of local sheep breeds. The animals studied belong to purebred and farms' own selection and tribal cards. The Kazakh Arkhar-Merino sheep breed was investigated on Kumtekey breeding farm, where the best sheep of this breed are kept. The Kazakh Arkhar-Merino is based on the interspecific hybridization of wild Arkhar rams with fine wool ewes of Novo-Caucasian Merino, Précoce, and Rambouillet breeds that occurred during 1934–1950. R-Kurty farm is also a breeding farm where the highly productive animals of the Kazakh Finewool breed are concentrated. Edilbay sheep breed is bred in Birlik breeding centre, where the purebred and highly productive sheep of this breed are kept. The Kazakh fat-tailed coarse wool sheep breed is the product of popular selection which has lasted many years. They are widely bred in many sheep farms all over Kazakhstan. However, highly productive animals typical of this breed are concentrated in Kabyl-Nur farm. Thus, four sheep breeds selected by us for molecular genetic studies are, firstly, the most common sheep breeds in Kazakhstan, which are well adapted to different climatic conditions of breeding and housing. Secondly, these breeds differ from each other in their origin and method of breeding, and thus they are excellent subjects for comparative molecular genetics research. Thirdly, the specification of breeding farms from which sheep were studied is extremely important, since we use the results of the molecular genetics study of these breeds in these farms to improve breeding work and speed up the selection process in order to create a highly productive breeding core in a short period of time. Since research started in 2010, the molecular database has grown, and the characterization of genetic diversity in farm animals has become particularly pertinent (Groeneveld et al., 2010).
Today, the FAO and the ISAG–FAO Advisory Group on Animal Genetic Diversity recommend specific sets of STR (short tandem repeat, also known as microsatellite) loci for genetic analyses, such as for horse, cattle, and pig breeds. Of the different types of molecular markers, STRs are suitable for studying genetic diversity because of their abundance – the large amount of allelic variation at each locus is highly polymorphic, their distribution throughout the genome is random, and inheritance is codominant (Rekha et al., 2016; Barcaccia et al., 2013; Putman et al., 2014). In addition, STRs are able to generate information for the planning of crossings and further selection of genotypes in genetic breeding programs (Faleiro et al., 2007; Crispim et al., 2014). The objective of the current research was to investigate 12 STR loci based on genetic diversity in sheep herds as well as the differentiation and relationship among the number of alleles and genetic links between Kazakh sheep breeds.
Blood samples were taken from five sheep populations: two of them were Edilbay-1, though of two various Edilbay-1 sheep herds, and three others were different sheep breeds in the categories Kazakh Finewool, Kazakh Arkhar-Merino, and Kazakh fat-tailed coarse wool. The 15 animals were chosen randomly from each population. Genomic DNA was extracted using a commercial kit (GeneJET Genomic DNA Purification Kit, ThermoFisher Scientific, USA). Both the quality and concentration of DNA were verified by spectrophotometric and agarose gel electrophoresis. In this study, 12 STR primers were used, and all of them are recommended by the International Society of Animal Genetics (ISAG, 2017). Amplification was carried out using a Tetrad 2 thermal cycler (Bio-Rad). Polymerase chain reaction (PCR) products were attached in the ABI310 Genetic Analyser, and GeneMapper software was used to determine fragment size. The number of alleles, effective number of alleles, polymorphism information content (PIC) values, observed and expected heterozygosities, Wright's F statistics per locus, pairwise population Fst values, and pairwise population matrix of Nei's standard genetic distance were calculated using GenAlex 6.5 and Excel microsatellite toolkit (version 3.1) software (Peakall and Smouse, 2012; Park 2008). The neighbour joining method of Saitou and Nei (1987) was used to construct a phylogenetic tree based on Nei's genetic distance in MEGA7 (Kumar et al., 2016). Factorial correspondence analysis (FCA) was investigated based on the individual multi-locus genotype using GENETIX version 4.03 (Belkhir et al., 1996).
All examined markers were polymorphic for all the populations examined. In total, 161 alleles were found in five native Kazakh sheep breeds based on 12 STR loci (Table 1). The highest number of alleles was 16 for markers INRA023 and CSRD247, whereas D5S2 showed the lowest number (8 alleles), and the average number of alleles was 13.4 per locus.
Table 1Genetic diversity analysis of sheep populations based on the 12 microsatellite markers. Number of alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), polymorphism information content (PIC), and F statistics (Fis, Fit, and Fst).
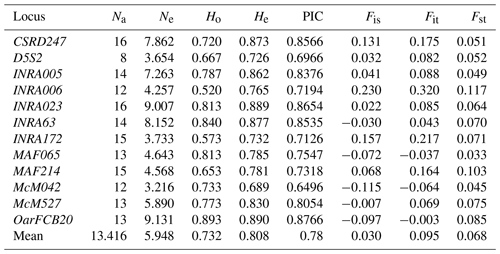
The effective number of alleles for each marker varied between 3.2 and 9.1, with a mean value of 5.9. The PIC values ranged from 0.65 to 0.88, with a mean of 0.78. Observed heterozygosity values varied from 0.52 to 0.89, with an average of 0.73, while the expected heterozygosity values ranged from 0.69 to 0.89 with a mean value of 0.81 (Table 1). Our study demonstrated a high genetic polymorphism in the investigated sheep breeds. These estimates are higher than those reported for other sheep breeds (Ferrando et al., 2014, in France and Spain; Salamon et al., 2014, in Croatia and Bosnia and Herzegovina; Al-Atiyat et al., 2015, in Australia; Gaouar et al., 2016, in Morocco).
Table 2Genetic diversity within the five sheep populations. Mean number of alleles (MNA), effective number of alleles (Ne), observed heterozygosity (Ho), and expected heterozygosity (He).
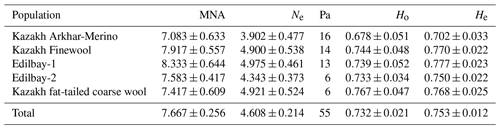
The average number of alleles, the effective number of alleles, and the expected and observed heterozygosity for each breed are shown in Table 2. The average number of alleles for the Edilbay-1 breed is 8.33. This analysis showed that there was no significant differentiation among the groups from the following populations: Kazakh Arkhar-Merino (7.08), Kazakh Finewool (7.91), Edilbay-2 (7.58), and Kazakh fat-tailed coarse wool (7.41). With the exception of Ne=3.90 in Kazakh Arkhar-Merino sheep, the effective number of alleles was larger than 4.0. In this study, the average value of observed heterozygosity was 0.68 for the Kazakh Arkhar-Merino population, and similar results were reported for Chinese and Mongolian breeds (Zhong et al., 2011). It is important to note that this is lower compared to the other four investigated sheep populations. At the same time, the expected heterozygosity value was more than 0.7 for all sheep groups. Values of the comparable means have been reported by Yilmaz et al. (2014).
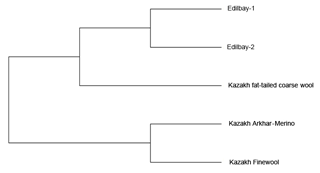
Figure 1The phylogenetic tree constructed from Nei's standard genetic distances among five sheep populations.
The highest Fis value was observed in marker INRA006, while the lowest Fis was recorded for locus McM527. The maximum and minimum Fit values were found in markers INRA006 and OarFCB20 respectively. Fst values ranged from 0.117 to 0.033, and the average values of Fis, Fit, and Fst were 0.030, 0.095, and 0.068 accordingly. According to the observed results of Fis, related mating occurs within each studied breed (Fis > 0). Obtained Fst values showed that the degree of genetic divergence is moderate between the sheep populations (Fst > 0.06). The Fit value was higher than zero, which indicates deficiency of heterozygosity. The highest genetic distance was found between Kazakh Arkhar-Merino and Edilbay-1 (0.469), while the smallest genetic distance was observed between Edilbay-1 and Edilbay-2 (0.217). Pairwise values of genetic differentiation, Fst, varied from 0.030 to 0.063.
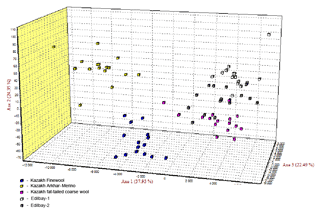
The neighbour joining tree for all samples was constructed using pairwise population matrix of Nei's genetic distances in order to represent the relationships between the sheep breeds (Fig. 1). Edilbay-1 and Edilbay-2 were initially classified as sub-clusters, which were further clustered into Kazakh fat-tailed coarse wool, whereas Kazakh Arkhar-Merino and Kazakh Finewool sheep breeds were grouped around the same node. Moreover, in the factorial correspondence analysis, the distinction of three clusters is illustrated by three axes showing variances of 37.93 %, 24.35 %, and 22.49 %, respectively (Fig. 2).
In this study, 12 STR (microsatellite) markers were used to evaluate genetic diversity in five populations. The analysis revealed no significant differences in the main genetic characteristics of the interbred population: number of alleles, effective number of alleles, and expected and observed heterozygosities. A high level of genetic diversity was observed in loci INRA023 and CSRD247, with a large number of alleles. According to Ozerov et al. (2008), the Edilbay sheep breed had the highest genetic diversity among the populations studied. Similarly, our analysis of the data showed that the greatest genetic diversity was detected in the Edilbay-1 sheep breed, with a mean value of alleles of 8.333±0.644, and these indicators did not differ much among the remaining populations (Table 2). The allele diversity for Edilbay-1 was similar to that of Arabian sheep studied with 12 microsatellite loci (Ahmed et al., 2018) but much higher than that reported for Jordan sheep breeds (Khaleel et al., 2018). The lowest genetic diversity among the breeds studied was expressed in Kazakh Arkhar-Merino, with an average value of seven alleles. The obtained allele diversity for Kazakh Arkhar-Merino was higher than the other studies (Sadeghi, 2018 and Khaleel et al., 2018), and this indicator is close to that described for the Algerian breeds (Gaouar et al., 2015). Moreover, in the present study the highest effective number of alleles was found for Edilbay-1(Ne=4.9), similar to values reported for Nellore sheep (Vani et al., 2017). The reason why the Edilbay-1 sheep breed maintains high genetic diversity is that currently in Birlik breeding centre there are 16 512 breeding sheep of the Edilbay breed, including 7500 ewes. In addition, farmers raise the sheep in different flocks by dividing them into three different groups according to the colour of their wool. Researchers have shown that animals with different colour wool are characterized by unequal productivity. For example, it has been proven that ewes with black wool have a higher wool yield by 7.5 %–11.8 %, a higher live weight by 2.2 %–6.9 %, and better slaughter qualities than sheep with red wool. The sheep with brown wool are characterized by the same high productivity indices. The fertility of females is not higher than 110 %–120 %. The milk content of the sheep is quite high. Furthermore, during the season of breeding, breeders exchange rams among flocks to maintain the genetic diversity.
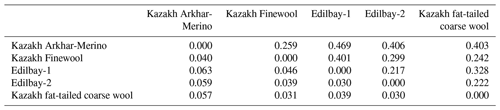
Table 3Pairwise population matrix of Nei's genetic distances (above the diagonal) and pairwise population Fst Values (below the diagonal).
The overall average of PIC value for the five populations was equal to 0.78, and all the markers were higher than 0.5, which means all the investigated STR loci were highly informative. This value was higher than that reported for Chinese sheep (PIC of 0.64; Guang-Xin et al., 2016). Also, PIC values showed that the most informative markers were OarFCB20 and INRA023. However, PIC values were lower than heterozygosities. According to Botstein et al. (1980), PIC must be always less than the expected heterozygosity. Aside from Kazakh Arkhar-Merino, in all examined populations, the average PIC value had a similar ranking. Consequently, these markers are suitable for studying the genetic diversity in Kazakh sheep breeds.
The fixation index was estimated on a per locus basis, and due to negative assortative mating, an excess of heterozygosity was found at markers MAF065, McM042, and OarFCB20.
In addition, to estimate the genetic variability of the studied sheep breeds, we calculated the expected and observed heterozygosity values. Except for MAF065 and McM042, in all loci the level of observed heterozygosity was lower than expected heterozygosity; therefore there were more homozygous individuals in the investigated flocks. As well as the mean expected heterozygosity in each population being higher than that of the heterozygotes, our results showed that heterozygotic deficiency was noted in all populations. Compared to the average heterozygosity value in the population, the observed heterozygotes fluctuated from 0.678±0.051 (Kazakh Arkhar-Merino) to 0.767±0.047 (Kazakh fat-tailed coarse wool), while the expected heterozygotes ranged from 0.702±0.033 (Kazakh Arkhar-Merino) to 0.777±0.023 (Edilbay-1). Meanwhile, the overall average values for this population were 0.732±0.021 and 0.753±0.012 (Table 2). These findings were similar to those reported in the literature (Sassi-Zaidy et al., 2014). The genetic variability obtained in these studies was similar to the results reached by Ozerov et al. (2008). By contrast, the level of genetic variability of the Kazakh Arkhar-Merino sheep breed is lower than those of Edilbay and Kazakh Finewool. In spite of this, this indicator of Kazakh Arkhar-Merino was still higher than those of some native Chinese sheep breeds (Guang-Xin et al., 2016) as well as eastern Adriatic and western Dinaric native sheep breeds (Salamon et al., 2014). The average mean of the observed heterozygosity was less than the mean of expected heterozygosity in the whole population; this could be due to selection against heterozygosity or inbreeding (Abdullah et al., 2013; Carmen et al., 2007). Thus, according to the comparison results of the investigated populations, the genetic variability in the Kazakh Arkhar-Merino population was identified as being lower than in the others.
Previously, a study was conducted by Ozerov et al. (2008) on four sheep breeds of Kazakhstan (Degeres Mutton-wool, Kazakh Arkhar-Merino, Kazakh Finewool, and Edilbaev) using 20 microsatellite loci. As a result, it was determined that all studied sheep breeds showed a high level of polymorphism in all 20 microsatellite loci and were in a state of genetic equilibrium according to the Hardy–Weinberg ratio. However, in our studies of the Kazakh Arkhar-Merino sheep breed, there were significant differences between the expected and observed heterozygosity (Ho=0.678 and He=0.702). In Ozerov (2008) et al., this index was in equilibrium (Ho=0.72 and He=0.71). This means that in the Kazakh Arkhar-Merino sheep breed population there is a deficiency of heterozygotes. This fact evidences that the degree of inbreeding of the Kazakh Arkhar-Merino sheep breed is quite high. In the current conditions of animal breeding, when private farm sheep breeding is widely practised in the country, breeding farms for one breed each have a small number of animals. Therefore, they practically do not exchange tribal animals among themselves. However, with a closed breeding system, inbreeding sooner or later reaches a dangerous level and leads to inbreeding depression (decreased productivity and reproductive qualities) and, finally, degeneration. Consequently, it is necessary to strive to limit inbreeding in conserved breeds and gene pool herds. The same situation exists for sheep of the Kazakh Arkhar-Merino breed from Kumtekey breeding farm. Based on the obtained genetic information on a high degree of inbreeding, animals from this farm will develop special breeding measures to preserve the valuable interspecific gene pool of animals of this unique breed and their rational use. We gave recommendations to Kumtekey breeding farm to reduce inbreeding urgently while maximizing the number of males used and reducing the number of lines with the highest intensity of interlinear pairing.
Furthermore, in order to analyse population differentiation and structure, Wright's F statistics of three indices for the overall population were used for each locus (Wright, 1965). Fis, the fixation coefficient of an individual within a subpopulation, shows a loss of heterozygosity in loci excluding INRA63, MAF065, McM042, McM527, and OarFCB20. The values of the inbreeding coefficient of an individual within the total population, Fit, ranged from 0.320 to −0.003. The fixation coefficient of the subpopulation within the whole population, Fst, indicates reduction of heterozygosity due to a limit of gene flow and genetic drift among the subpopulation. This result indicates a moderate degree of genetic divergence among subpopulations or breeds of 6.8 % overall. At the same time, the degree of genetic divergence of a subpopulation within the total population amounted to 93.2 %. A similar result was found by Gaouar et al. (2014), (Fst=6.1 %), who studied the genetic admixture of North African ovine breeds based on six STR loci (Gaouar et al., 2014). The Fst value was considerably higher than both the Fst values previously reported by Ozerov et al. (2008) and that of Algerian sheep breeds (Gaouar et al., 2015; Abdelkader et al., 2017).
In the present study, we used pairwise population Fst values to provide a measure of the genetic differentiation among sheep breeds (Table 3). The proportion of genetic divergence indicated that moderate differentiation was observed between Kazakh Arkhar-Merino and Edilbay-1 (0.063), Kazakh Arkhar-Merino and Edilbay-2 (0.059), and Kazakh Arkhar-Merino and Kazakh fat-tailed coarse wool (0.057), with a mean of Fst > 0.05. These obtained differentiation parameters are generally comparable with Fst values of the fat-tailed Barbarine sheep breed (Sassi-Zaidy et al., 2014). The other investigated sheep population for pairwise genetic differentiation parameters showed values < 0.05, which, according to Hartl (1980), indicates a low differentiation among the population. Further, significant genetic differentiation observed ranged from 0.031 to 0.046. Mahmoud et al. (2017) also found significant differentiation between Sawakni, Berberi, and Najdi sheep breeds. However, the lowest Fst coefficient is found between Edilbay-1 and Edilbay-2 since Edilbay-1 and Edilbay-2 are classified as one breed. In order to enhance breed character and to attempt an increase in the status of Edilbay-2, the farmers are buying males from the Edilbay-1 breed, for the reason that the Edilbay-1 breed is a livestock breeding farm with the best quality in Kazakhstan. Consequently, it is clear that migration is going on here. Migration has a great effect on the reduction of genetic differentiation among the population (Laval et al., 2000).
Nei's genetic distances (Nei, 1972) between the five populations of sheep were calculated using 12 STR loci (Table 3), which varied from 0.469 to 0.217. The results of genetic distance of the present study were lower than the findings of Bai et al. (2015), who reported that the genetic distance ranged from 0.21 to 0.62 for Chinese indigenous sheep breeds, which was higher than that of Egyptian sheep breeds (Rushdi et al., 2015). The values of genetic distances showed that the investigated sheep populations are characterized as having a high range of variability of the allele pool, presence or absence of certain alleles, and differences in frequency of occurrence alleles. The largest genetic distance was observed between Kazakh Arkhar-Merino and Edilbay-1 (0.469). These two breeds were absolutely different from each other originally: by phenotype, according to the different branches of stock breeding and geographical places. Kazakh Arkhar-Merino is a meat-woolly breed with fine wool. Kazakh Arkhar-Merino is well adapted to breeding at high altitudes, and these sheep differ favourably from other breeds in conditions of mountain pasture. In contrast, the Edilbay-1 sheep breed is classified as the coarse wool sheep from the meat-fatty category. They are well adapted to the severe desert and semi-desert conditions of Kazakhstan. Also, Kazakh Arkhar-Merino is geographically most distant from Edilbay-1. In comparison with other populations, a closer relationship was found between Kazakh Arkhar-Merino and Kazakh Finewool (0.25), which could be attributed to the low geographical distance between these two populations. The same result has been found between Kermani and Lori-Bakhtiari Pakistani sheep (0.25), which both breed in Iran (Vajed Ebrahimi et al., 2017). Both of them (Kazakh Arkhar-Merino and Kazakh Finewool) refer to the meat and wool types of sheep breeds. Kazakh fat-tailed coarse wool and Edilbay-2 had less similarity to the Kazakh Finewool than Edilbay-1.
Further, to assess the genetic relationships among the population, a phylogenetic tree was constructed using the neighbour joining method (Saitou and Nei, 1987) based on Nei's genetic distance. The results of the phylogenetic tree indicated that the Edilbay-1 and Edilbay-2 sheep breeds were clustered into Kazakh fat-tailed coarse wool, which are coarse wool breeds and have a common origin. However, Kazakh Finewool and Kazakh Arkhar-Merino were grouped in the same node, both being fine wool sheep, as these breeds have the same ancestral background. Likewise, these two breeds were categorized together on the same branch of the phylogenetic tree in the previous study (Ozerov et al., 2008).
In addition, factorial correspondence analysis demonstrated that Kazakh Arkhar-Merino and Kazakh Finewool were isolated from the other studied populations due to the high level of differentiation and no sharing of alleles. Edilbay-2 is an admixture of both Edilbay-1 and Kazakh fat-tailed coarse wool. According to the FCA findings, this is connected with historical origin of these Kazakh sheep breeds.
In this study, within and among herds, the genetic diversity of five Kazakh sheep populations was assessed using 12 microsatellite markers. Based on our results, all five populations examined show high genetic diversity through a high effective number of alleles, a large mean number of alleles, high PIC values, and 12 completely polymorphic tested microsatellites. Moderate differentiation was found between Kazakh Arkhar-Merino and Edilbay-1, whereas differentiation between Edilbay-1 and Edilbay-2 was lower. Therefore, the evaluation of the results of Nei's genetic distance, neighbour joining, and FCA agreed with the historical origin of animals. As our study showed, although 10 years have passed since the research by Ozerov et al. (2008), it has been discovered that genetic diversity remains in sheep breeds other than Kazakh Arkhar-Merino. The main reasons for this fact are as follows. Scientists and livestock breeders have always been working on the improvement of these breeds. The advantages of this research also include the ability of the breeds to transmit all useful economic traits to their offspring. Breeding work is carried out mainly along the lines using several herds. Of course, not only the best males, but also elite queens, are selected to replenish livestock. Lines of sheep are created according to some outstanding qualities – precocity, weight, size of fat tail, and quality of wool. The work consists mainly of mating animals with distant degrees of kinship in the line. In addition, based on the data obtained, it is possible to recommend the Kazakh Arkhar-Merino breed for the selection against homozygous individuals. Further, the results achieved on STR loci are proposed to be used to control and conserve the genetic diversity of native sheep breeds.
The materials used the current study are available from the corresponding author on reasonable request.
KD and ZO participated in the collection of all materials and conducted all the experiments. KD and AM wrote the paper. RZh, BB, NS, and BM participated in the editing of the paper. All authors read and approved the final paper.
The authors declare that they have no conflict of interest.
The protocol for the use of materials was agreed with the Commission on Bioethics of the Kazakh National Agrarian University, Almaty, Kazakhstan.
The authors thank Gulnur Zhunussova for helping in all experiments and for technical support.
This research has been supported by the Ministry of Education and Science of the Republic of Kazakhstan (grant no. 541).
This paper was edited by Steffen Maak and reviewed by Semir Bechir Suheil Gaouar and one anonymous referee.
Abdelkader, A. A., Ata, N., Benyoucef, M. T., Djaout, A., Azzi, N., Yilmaz, O., Cemal, I., and Gaouar, S. B. S.: Newgenetic identification and characterisation of 12 Algerian sheep breeds by microsatellite markers, Ital. J. Anim. Sci., 1, 38–48, https://doi.org/10.1080/1828051X.2017.1335182, 2017.
Abdullah, G., Samir, M., Manal, E., Ashraf, A., Mohamd, E., Keiichiro, K., and Kazuyoshi, H.: Genetic diversity and distances of three Egyptian local sheep breeds using microsatellite markers, Research in Zoology, 3, 1–9, https://doi.org/10.14202/vetworld.2018.778-781, 2013.
Ahmed, H. M., Faisal, M. A., Ahmed, R., Khalid, M. A., Mohammad, A. F., and Osama, B. M.: Genetic variability of sheep populations of Saudi Arabia using microsatellite markers, Indian J. Anim. Res., 775, 1–4, 2018.
Al-Atiyat, R. M.: The power of 28 microsatellite markers for parentage testing in sheep, Electron. J. Biotechn., 18, 116–121, 2015.
Bai, J. Y., Yang, Y. B., Wang, Y. Q., Zhang, X. H., and Pang, Y. Z.: Polymorphism analysis of three Chinese indigenous sheep breeds by microsatellite markers, Indian J. Anim. Res., 49, 585–590, https://doi.org/10.18805/ijar.5566, 2015.
Barcaccia, G., Felicetti, M., Galla, G., Capomaccio, S., Cappelli, K., Albertini, E., Buttazzoni, L., Pieramati, C., Silvestrelli, M., and Supplizi, A. V.: Molecular analysis of genetic diversity population structure and inbreeding level of the Italian Lipizzan horse, Livest Sci., 151, 124–133, https://doi.org/10.1016/j.livsci.2012.11.022, 2013.
Belkhir, K., Borsa, P., Goudet, J., Chikhi, L., and Bonhomme, F.: GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations, in: Laboratoire Génome Populations, Interactions CNRS UMR 5000, Université de Montpellier II, Montpellier, France, 1996.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, 1980.
Carmen, M.: Genetic characterization of indigenous goat populations of Mozambique, Master thesis, Department of Production Animal Studies in Faculty of Veterinary Science, University of Pretoria, South Africa, 2007.
Crispim, B. A., Seno, L. O., Egito, A. A., Junior, F. M. V., and Grisolia, A. B.: Application of microsatellite markers for breeding and genetic conservation of herds of Pantaneiro sheep, Electron. J. Biotechn., 17, 317–321, https://doi.org/10.1016/j.ejbt.2014.09.007, 2014.
Faleiro, F. G.: Marcadores genético-moleculares aplicados a programas de conser-vação e uso de recursos genéticos, Planaltina-DF, Embrapa Cerrados, 2007.
Ferrando, A., Goyache, F., Parés, P. M., Carrión, C., Miró, J., and Jordana, J.: Genetic relationships between six eastern Pyrenean sheep breeds assessed using microsatellites, Span. J. Agric. Res., 12, 1029–1037, https://doi.org/10.5424/sjar/2014124-6173, 2014.
Gaouar, S. B. S., Kdidi, S., Tabet Aouel, N., Aït-Yahia, R., Boushaba, N., Aouissat, M., Dhimi, L., Yahyaoui, M. H., and Saidi-Mehtar, N.: Genetic admixture of North-African ovine breeds as revealed by microsatellite loci, Livest. Res. Rural. Dev., 26, 1–11, http://www.lrrd.org/lrrd26/7/gaou26118.htm(last access: 25 Mat 2019), 2014.
Gaouar, S. B. S., Da Silva, A., Ciani, E., Kdidi, S., Aouissat, M., Dhimi, L., Lafri, M., Maftah, A., and Mehtar, N.: Admixture and local breed marginalization threaten Algerian sheep diversity, PLoS ONE, 10, e0122667, https://doi.org/10.1371/journal.pone.0122667, 2015.
Gaouar, S. B. S., Kdidi, S., and Ouragh, L.: Estimating population structure and genetic diversity of five Moroccan sheep breeds by microsatellite markers, Small Ruminant. Res., 144, 23–27, 2016.
Groeneveld, L. F., Lenstra, J. A., Eding, H., Toro, M. A., Scherf, B., Pilling, D., Negrini, R., Finlay, E. K., Jianlin, H., Groeneveld, E., and Weigend, S.: Genetic diversity in farm animals – a review, Anim. Genet., 41, 1–26, https://doi.org/10.1111/j.1365-2052.2010.02038.x, 2010.
Guang-Xin, E., Tao, Z., Yue-Hui, M., Hui-Jiang, G., Jian-Ning, H., and Nan, L.: Conservation genetics in Chinese sheep: diversity of fourteen indigenous sheep (Ovis aries) using microsatellite markers, Ecol. Evol., 6, 810–817, https://doi.org/10.1002/ece3.1891, 2016.
Hartl, D.: Principles of population genetics. Sunderland, MA, Sinauer Association Inc, Massachusset, USA, 164–165, 1980.
ISAG: 36th International Society for Animal Genetics Conference, 16–21 July, Dublin, Ireland, available at: https://www.isag.us/Docs/AppGenSheepGoat2017.pdf (last access: 28 May 2019), 2017.
Khaleel, I. J., Mustafa, M. A., Zuhair, B. I., Abdel, M. B. Y., and Ibrahem, A. S.: Genetic diversity and population structure of local and exotic sheep breeds in Jordan using microsatellites markers, Veterinary World, 11, 778–781, 2018.
Kumar, S., Stecher, G., and Tamura, K.: MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 33, 1870–1874, https://doi.org/10.1093/molbev/msw054, 2016.
Laval, G., Iannuccelli, N., Legault, C., Milan, D., Groenen, M. A., Giuffra, E., Andersson, L., Nissen, P. H., Jørgensen, C. B., Beeckmann, P., Geldermann, H., Foulley, J. L., Chevalet, C., and Ollivier, L.: Genetic diversity of eleven European pig breeds, Genet. Sel. Evol., 32, 187–203, https://doi.org/10.1051/gse:2000113, 2000.
Mahmoud, A. H., Swelum, A., Farah, M. A., Alanazi, K., Rady, A., Salah, M., Amor, N., and Mohammed, O. B.: Genetic diversity among Sawakni, Berberi and Najdi sheep breeds in Saudi Arabia using microsatellites markers, Afr. J. Biotechnol., 16, 171–178, https://doi.org/10.5897/AJB2016.15785, 2017.
Nei, M.: Genetic distance between populations, Am. Nat., 106, 283–292, 1972.
Ozerov, M. Y., Marzanov, N. S., Tapio, M., Burabaev, A. A., Marzanova, L. K., Amerkhanov, K. A., and Kantanen, Yu: Genetic features of Kazakh sheep breeds according to microsatellites, Russian Agricultural Sciences, 34, 45–48, https://doi.org/10.3103/S1068367408010175, 2008.
Park, S. D. E.: The Excel microsatellite Toolkit (version 3.1), Animal Genomics Laboratory, University College, Dublin, Ireland, 2001, available at: http://animalgenomics.ucd.ie/sdepark/ms-toolkit/, last access: April 2008.
Peakall, R. and Smouse, P.: GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research – an update, Bioinformatics, 28, 2537–2539, https://doi.org/10.1093/bioinformatics/bts460, 2012.
Putman, A. I. and Carbone, I.: Challenges in analysis and interpretation of microsatellite data for population genetic studies, Ecol. Evol., 4, 4399–428, https://doi.org/10.1002/ece3.1305, 2014.
Rekha, S., Brijesh, K., Reena, A., Sonika, A., Mishra, A. K., and Tantia, M. S.: Genetic diversity estimates point to immediate efforts for conserving the endangered Tibetan sheep of India, Meta Gene, 8, 14–20, https://doi.org/10.1016/j.mgene.2016.01.002, 2016.
Rushdi, H. E. and Sabry, A. M.: Analysis of the genetic diversity of three Egyptian sheep breeds using microsatellite markers, International Journal of Advanced Research, 3, 112–120, 2015.
Sadeghi, B.: Genetic Diversity of Tandureh Mouflon Population, Acta Vet. Eurasia, 44, 20–25, 2018.
Saitou, N. and Nei, M.: The neighbor-joining method: A new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 4, 406–425, https://doi.org/10.1093/oxfordjournals.molbev.a040454, 1987.
Salamon, D., Gutierrez-Gil, B., Arranz, J. J., Barreta, J., Batinic, V., and Dzidic, A.: Genetic diversity and differentiation of 12 eastern Adriatic and western Dinaric native sheep breeds using microsatellites, Animal, 8, 200–207, https://doi.org/10.1017/S1751731113002243, 2014.
Sassi-Zaidy, Y. B., Maretto, F., Zanetti, E., Hajji, G. M., Charfi-Cheikrouha, F., and Cassandro, M.: Genetic structure and variability within and among populations of the fat-tailed Barbarine sheep breed using microsatellites markers, Afr. J. Biotechnol., 13, 44–54, https://doi.org/10.5897/AJB2013.13363, 2014.
Vajed Ebrahimi, M. T., Mohammadabadi, M., and Esmailizadeh, A.: Using microsatellite markers to analyze genetic diversity in 14 sheep types in Iran, Arch. Anim. Breed., 60, 183–189, https://doi.org/10.5194/aab-60-183-2017, 2017.
Vani, S., Punyakumari, B., Gnanaprakash, M., and Rao, V. H.: Microsatellite marker analysis in Nellore breed of sheep, International Journal of Science, Environment and Technology, 6, 702–707, 2017.
Wright, S.: The interpretation of population structure by F-statistics with special regard to systems of mating, Evolution, 19, 395–420, https://doi.org/10.1111/j.1558-5646.1965.tb01731.x, 1965.
Yilmaz, O., Cemal, I., and Karaca, O.: Genetic diversity in nine native Turkish sheep breeds based on microsatellite analysis, Ani. Genet., 45, 604–608, https://doi.org/10.1111/age.12173, 2014.
Zhong, T., Han, J. L., Guo, J., Zhao, Q. J., Fu, B. L., Pu, Y. B., He, X. H., Jeon, J. T., Guan, W. J., and Ma Y. H.: Tracing genetic differentiation of Chinese Mongolian sheep using microsatellites, Anim. Genet., 42, 563–565, https://doi.org/10.1111/j.1365-2052.2011.02181.x, 2011.