the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Genetic variations in the sheep SIRT7 gene and their correlation with body size traits
Hongwei Xu
Xiaoyu Zhang
Rongxin Zang
Yong Cai
Xin Cao
Jutian Yang
Jie Li
Xianyong Lan
Jianping Wu
As a nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase and ADP ribosyl transferase, the silent information regulator 7 (Sirtuin 7, SIRT7) plays a crucial role in regulating the differentiation of adipocytes and myoblasts, lipid metabolism, glucose metabolism, and cellular growth in mammals. It has been hypothesized that SIRT7 affects growth traits in animals; therefore, in this study, the potential insertion/deletion (indel) of genetic variations within the ovine SIRT7 gene and their correlation with sheep growth traits were explored. A total of 709 individuals from five Chinese and Mongolian sheep breeds were analyzed. Two novel indel loci of the sheep SIRT7 gene were detected and were named 5′ promoter region-insertion-7 bp (5′ promoter region-7 bp) and 3′ UTR-insertion-17 bp (3′ UTR-17 bp), respectively. In all of the sheep breeds, frequencies of the 5′ promoter region-7 bp mutation were low, whereas mutations of 3′ UTR-17 bp were high in Tong sheep and Lanzhou fat-tail sheep (LFTS). Furthermore, both indel polymorphisms had significant associations with different growth characteristics (P<0.05). Among these associations, the 3′ UTR-17 bp was highly correlated with rump width in small-tail Han sheep (STHS, rams; P<0.01), and individuals with the ID genotype had better chest depth values than those with the II genotype. In this paper, two novel indels within the sheep SIRT7 gene were identified, and genetic diversity and its effects on body size traits were explored. These findings will potentially provide useful DNA markers for the improvement of economic traits in sheep genetic breeding.
- Article
(3513 KB) - Full-text XML
- BibTeX
- EndNote
Silent information regulator 2 (sirtuins2, SIRT2) proteins, which belonging to the class III nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylases and ADP ribosyl transferases, are connected to metabolism and life span regulation in lower organisms and are highly conservative from prokaryotes to eukaryotes (O'Callaghan and Vassilopoulos, 2017). In particular, the mammalian sirtuin proteins have been reported to participate in the regulation of energy metabolism (e.g., sugar and lipid metabolism) (Li and Kazgan, 2011; Ye et al., 2017) and stress (Blank and Grummt, 2017), as well as the maintenance of genomic stability (Bosch-Presegué and Vaquero, 2014), tumor development (Roth and Chen, 2014), cell proliferation, senescence, and apoptosis (O'Callaghan and Vassilopoulos, 2017).
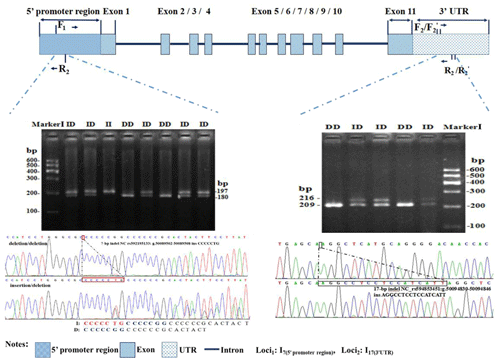
Figure 1Electrophoresis diagram and sequence of 3′ UTR-insertion-17 bp loci and 5′ promoter region-insertion-7 bp within the ovine SIRT7 gene.
Recently, SIRT genes have been shown to regulate the differentiation of adipocytes and myoblasts (Cioffi et al., 2015). Additionally, an increasing number of studies have revealed that the genetic diversity of SIRT genes is related to the economic traits of livestock. For instance, single nucleotide polymorphisms (SNPs) of the bovine SIRT1 gene have been shown to be significantly correlated with carcass traits (such as dressing percentage, meat percentage, and carcass weight) of Chinese Luxi cattle (Liu et al., 2017). Moreover, g.13915A>G within the SIRT4 gene significantly influences body size traits (body length, chest depth, rump length, and chest circumference) in Chinese Qinchuan cattle (Gui et al., 2016). There are seven members in the mammalian sirtuin protein family (SIRT1–SIRT7) that have different subcellular localizations. As the only member that is predominantly localized to the nucleolus (Michishita et al., 2005), SIRT7 has been reported to be involved in cellular growth and metabolism. At the cellular level, SIRT7 participates in the physiological processes of the cell stress response, proliferation, apoptosis, aging, and so on (Shin et al., 2016; Blank and Grummt, 2017). At the individual level, SIRT7 can control the progress of glycolysis (Jiang et al., 2017; Ye et al., 2017) and hepatic lipid metabolism by regulating the ubiquitin–proteasome pathway (Yoshizawa et al., 2014; Tang et al., 2015; Ye et al., 2017; Yamagata and Yoshizawa, 2018). Previous studies have revealed that SIRT7 antagonizes TGF-β signaling to inhibit breast cancer cell metastasis (Tang et al., 2017) and promotes glioma proliferation and invasion via the activation of the ERK/STAT3 signaling pathway (Mu et al., 2019). Notably, two synonymous SNP mutations of bovine SIRT7 have been confirmed to be significantly associated with the body length, hip length, back fat thickness, and chest circumference of Qinchuan cattle (Gui et al., 2016). In contrast, there are few related reports regarding ovine SIRT7.
The ovine SIRT7 gene is located on chromosome 11, and includes 11 exons. The molecular and biological properties of the SIRT7 gene make it a potential candidate gene to affect body size traits in animals; however, the associations between the body size traits of sheep and mutations in the SIRT7 gene are poorly explored and researched, especially regarding the insertion/deletion (indel) variants. Compared with other molecular markers (including SNPs), indel variants have superiority in terms of detection efficiency (Yang et al., 2016). Therefore, to explore the indel polymorphisms of the ovine SIRT7 gene, five representative sheep breeds were studied. The Sartuul sheep (SS) is a vital wool/meat dual purpose domestic sheep breed in Mongolia, whereas the Lanzhou fat-tail sheep (LFTS), the small-tail Han sheep (STHS), the Tong sheep (TS), the and Hu sheep (HS) are representative indigenous sheep breeds in China (Li et al., 2018a, b). The objectives of this study were to investigate the novel indel polymorphisms within the ovine SIRT7 gene in the five abovementioned sheep breeds and explore the correlations of these novel indel polymorphisms with ovine body size traits. The results of this study could potentially accelerate the progress of sheep breeding.
2.1 Animal welfare statement
In this study, all experimental processing was fully consistent with the animal welfare guidelines, laws and policies of Northwest A&F University.
2.2 Collection of DNA samples and data
In total, 737 same-aged sheep from five different breeds were used: four indigenous Chinese breeds, STHS (n=187), TS (n=165), LFTS (n=58), and HS (n=189); and one Mongolian native breed, SS (n=138) (Li et al., 2018a, b; Ma et al., 2018a, b). Using the same standard, the body traits of all of the sheep were measured (Zhao et al., 2017), including the cannon circumference (CaC), body weight (BW), and chest depth (ChD), among others. Thereafter, related growth trait indices, such as the chest width index (ChWI), were calculated according to the method from Lan et al. (2007).
2.3 Construction of genomic DNA pool
Using the high salt extraction method, the sheep genomic DNA was isolated from ear tissue which was preserved in 70 % alcohol at −80 ∘C (Lan et al., 2007). The samples were then assayed using a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA); the concentration of all of the diluted DNA samples was 10 ng µL−1 (Li et al., 2018b). Additionally, 50 samples of DNA were randomly selected and mixed to detect potential indel loci within the sheep SIRT7 gene (Yang et al., 2017).
2.4 Mutation loci amplification and production sequencing
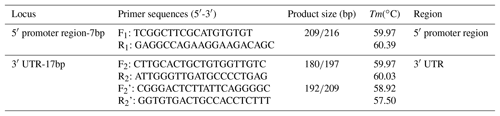
On the basis of the Ensembl database (https://asia.ensembl.org/, last access: 28 March 2019), two indel loci were selected in the sheep SIRT7 gene, one was located in the 5′ promoter region and the other was located in the 3′ UTR (Fig. 1). Referencing the gene sequence of the sheep SIRT7 gene (GenBank no: NC_019468.2), three pairs of amplification primers were designed using the Primer-BLAST tool in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome, last access: 28 March 2019) (Table 1) and were synthesized by Tsingke Biotech Company (Xi'an, China).
The amplification reaction was performed in a 13 µL volume system, including 1 µL of genomic DNA (10 ng µL−1), 6.5 µL of 2× EasyTaq PCR SuperMix (+dye), 0.5 µL of each primer, and 4.5 µL of ddH2O. The PCR amplification procedure was touch-down PCR (TD-PCR) using the steps described in previous studies (Yang et al., 2016; Xu et al., 2015; Li et al., 2018a). The amplification products were detected using 3.5 % agarose gel electrophoresis; the amplification products of each pair of primers were only sequenced if they were different (Zhang et al., 2015; Cui et al., 2018).
Table 2Genetic diversity parameters of novel polymorphisms of the ovine SIRT7 gene.
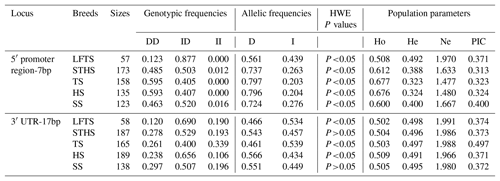
Note: HWE – Hardy–Weinberg equilibrium; Ho – homozygosity; He – heterozygosity; Ne – effective allele numbers; PIC – polymorphism information content.
2.5 Statistical analysis
The mutation sequences of the two new indel loci were verified by contrasting and analyzing the sequences using BioEdit software (BioEdit, USA). Frequencies of the genotypes and alleles of the sheep SIRT7 gene between different breeds were directly calculated using a Chi-square (χ2) test. Based on the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx, last access: 28 March 2019), the polymorphism information content (PIC) was calculated (Li et al., 2018a). In addition, the values of the r2 test and D' test were also calculated using the SHEsis program (http://analysis.bio-x.cn, last access: 28 March 2019) to analyze the linkage disequilibrium of the populations of the two mutation loci (Shi et al., 2005; Li et al., 2009, 2018a; Wang et al., 2017). Furthermore, using an analysis of variance (ANOVA) and independent samples t tests in SPSS software (version 23.0; IBM Corp, USA), the relationships between the indels of SIRT7 and the body size traits (e.g., chest width) in different breeds were determined (Li et al., 2018b). The genotypes were considered to be the independent variables and the growth traits were the dependent variables (Li et al., 2018a). If the data did not show homogeneity of variances and a normal distribution, a non-parametric (Kruskal–Wallis) test was used (Cui et al., 2018; Wang et al., 2019). The results were regarded as statistically significant when P<0.05, and all statistical tests were bilateral.
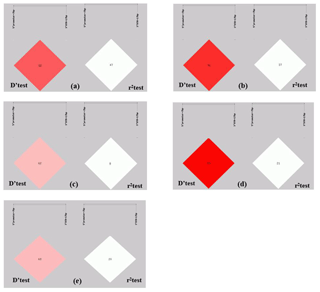
Figure 2Linkage equilibrium test of two pairs of alleles within the ovine SIRT7 gene in different populations: (a) Lanzhou fat-tail sheep, (b) small-tail Han sheep, (c) Tong sheep, (d) Hu sheep, and (e) Sartuul sheep. Note: the 5′ promoter region-insertion-7 bp (loci1) and 3′ UTR-insertion-17 bp loci (loci2) were chosen for haplotype analysis.
3.1 Genotyping of individuals
Two novel indel loci within the ovine SIRT7 gene, namely the 5′ promoter region-insertion-7 bp (5′ promoter region-7 bp) and 3′ UTR-insertion-17 bp (3′ UTR-17 bp), were genotyped and identified via gel agarose electrophoresis (3.5 %) and DNA sequencing (Fig. 1).
Polymorphisms of each indel had three different genotypes. For 5′ promoter region-7 bp (Fig. 1), genotype II (insertion/insertion) showed one band of 209 bp, genotype DD (deletion/deletion) exhibited one band of 216 bp, whereas the heterozygote genotype ID (insertion/deletion) exhibited two bands (216 and 209 bp). For 3′ UTR-17 bp (Fig. 1), three different bands were also exhibited: 180 bp, 197 bp, and 197/180 bp. After contrasting and analyzing mutant sequences, the two new indel loci were verified. The insertion sequences were CCCCCTG for 5′ promoter region-7 bp and AGGCCTCCTCCATCATT for 3′ UTR-17 bp, which were in agreement with the information in the Ensembl database.
3.2 Genetic parameters calculation
In Table 2, the frequencies of population parameters, genotypes, and alleles for the two indel loci in the five breeds tested are shown. For 5′ promoter region-7 bp, the DD and ID genotypes had higher frequencies than the II genotype in all of the breeds analyzed, and the D allele had a higher frequency than the I allele. For 3′ UTR-17 bp, the five breeds had a different dominant allelic frequency. In STHS, HS, and SS, the D allele had a higher frequency than the I allele, whereas the opposite was found in the LFTS and TS breeds. Additionally, the results of the population parameters demonstrated that these two indel markers displayed moderate polymorphism, and the PIC among them ranged from 0.313 to 0.497 in all of the breeds studied. Moreover, the 5′ promoter region-7 bp locus in all breeds tested, and the 3′ UTR-17 bp locus in the LFTS, TS, and HS breeds, did not conform to the Hardy–Weinberg equilibrium (HWE; P<0.05).
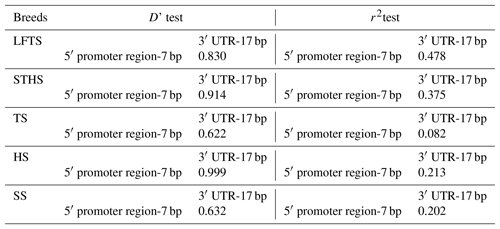
The r2 test showed that the 5′ promoter region-7 bp and the 3′ UTR-17 bp loci were at linkage equilibrium in all of the sheep groups tested (Table 3, Fig. 2). However, the D' test showed the opposite: in the SS and TS breeds, the D' test values showed that the two indel loci were in the weak linkage disequilibrium. Furthermore, the haplotype analysis revealed that there were four haplotypes and that “” had the highest incidence in all of the sheep groups detected (Fig. 3).
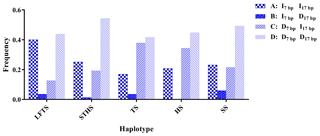
Figure 3Haplotype frequency of the 5′ promoter region-insertion-7 bp and 3′ UTR-insertion-17 bp loci of the ovine SIRT7 gene in different breeds. Note: LFTS – Lanzhou fat-tail sheep; STHS – small-tail Han sheep; TS – Tong sheep; HS – Hu sheep; and SS – Sartuul sheep.
Table 4Association of the novel indel of the ovine SIRT7 gene and growth traits in different breeds (LSMa ± SE).
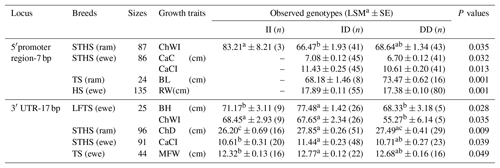
Note: ChWI – chest width index; CaC – cannon circumference; CaCI – cannon circumference index; BL– body length; RW – rump width; BH – body height; ChD – chest depth; and MFW – maximum forehead width. The different letters (a and b, or a and c) beside values within the same row signify significance at the P<0.05 and P<0.01 level, respectively.
3.3 Association of the indel polymorphisms and body size traits
The correlation of the two abovementioned novel indels and sheep body size traits were investigated to explore whether the polymorphisms of the ovine SIRT7 gene were related to sheep growth (Table 4). As shown in Table 4, both indel loci displayed significant relationships with the different sheep body size traits. The polymorphisms of the 5′ promoter region-7 bp were significantly associated with the chest width index (ChWI) in STHS (rams), the cannon circumference and cannon circumference index (CaC and CaCI, respectively) in STHS (ewes), the body length and chest depth (BL and ChD, respectively) in TS (rams), and the rump width in HS (ewes). Furthermore, mutations of the 3′ UTR-17 bp showed a significant relationship with the body height and ChWI in LFTS (ewes), the ChD in STHS (rams), the CaCI in STHS (ewes), and the maximum forehead width in TS (ewes). In particular, in STHS (rams), the effects of the 3′ UTR-17 bp on the chest depth was highly significant (P=0.009).
Studies of SIRT genes tend to focus on the regulation of the physiological processes of cell stress responses, proliferation, apoptosis, aging, metabolism, and especially on the development of tumors (Li and Kazgan, 2011; Roth and Chen, 2014; O'Callaghan and Vassilopoulos., 2017; Ye et al., 2017). Although, in recent years, SIRT genes have also been shown to regulate the differentiation of adipocytes and myoblasts (Cioffi et al., 2015). Furthermore, the genetic mutations of SIRT genes have revealed their effects on the production traits of livestock (Gui et al., 2016; Liu et al., 2017). However, existing studies on the polymorphism of SIRT genes have mostly focused on SNP or haplotype mutations in cattle, whereas the study of polymorphisms of ovine SIRT genes is lacking. Thus, this study is the first to identify genetic diversities in the ovine SIRT7 gene and explore the effects of these novel indel polymorphisms in representative Chinese and Mongolian sheep breeds. In this paper, two novel indels, both located in the untranslated region of the ovine SIRT7 gene, were identified. For the 5′ promoter region-7 bp, the D allele had a higher frequency than the mutant allele (I), whereas the sheep breeds tested had different dominant allelic frequencies in 3′ UTR-17 bp. Moreover, the indel polymorphisms detected did not conform to the HWE in several of the breeds tested (e.g., 5′ promoter region-7 bp in STHS and 3′ UTR-17 bp in LFTS). Migration, crossbreeding, small sample numbers, and long-term artificial selection are suspected to be the factors contributing to the deviation from the HWE (Li et al., 2018a).
In mammals, polymorphisms of the SIRT7 gene are involved in human longevity (Donlon et al., 2018), high-altitude adaptation (Y. Li et al., 2014), and Parkinson's disease (Jesús et al., 2013). Because of the key role of SIRT7 in regulating lipid metabolism (Yoshizawa et al., 2014; Tang et al., 2015), glucose metabolism (Jiang et al., 2017; Ye et al., 2017), and cell growth or differentiation, the relationship between the polymorphisms tested and sheep growth traits were also analyzed. Through the association analysis, all indel loci detected were found to be significantly associated with sheep body size traits, and several associations were highly significant. For instance, in 3′ UTR-17 bp, STHS rams with the ID genotype had a greater ChD than individuals with the II genotype.
Previous studies have revealed that the mutation polymorphisms in the UTR could affect the expression of the gene by affecting the functions of elements of DNA or via microRNA-mediated post-transcriptional regulation mechanisms (Krawczak et al., 1992; Komar, 2007; G. Li et al., 2014; Tak and Farnham, 2015). Nevertheless, in this study, the location of 3′ UTR-17 bp was approximately 100 bp upstream of the seed sequence of microRNA. Furthermore, the interactions between the target gene and its adjacent genes were also speculated to contribute to this association (Li et al., 2018a, b). MAF bZIP transcription factor G (MAFG), situated in the upstream 3 kb of SIRT7 was considered to be a transcriptional repressor whose over expression could inhibit the synthesis and metabolism of bile acid (de Aguiar Vallim et al., 2015; Katsuoka and Yamamoto, 2016). Moreover, Phosphate cytidylyltransferase 2, ethanolamine (PCYT2), located downstream 1 kb of SIRT7, could regulate muscle cell differentiation (Zhu et al., 2009). The mutation of UTR in the ovine SIRT7 gene may influence the expression of the adjacent genes, thereby affecting the growth traits of sheep. However, the specific regulatory mechanisms still require further study.
In summary, two novel indel loci within the ovine SIRT7 gene, 5′ promoter region-insertion-7 bp and 3′ UTR-insertion-17 bp, were detected in sheep. Moreover, the results of correlation analyses demonstrated that the indel loci were significantly associated with sheep growth traits, which indicated that the novel indel loci could potentially be useful DNA markers for the genetic improvement of economic traits in sheep breeding.
Data sets are available upon request from the corresponding authors.
HZ and XZ contributed equally to this work. JW and XL designed the study. RZ, YC, XC, and JY collected the samples. XZ and JL performed the experiments. This paper was written by HX. JW corrected the paper and gave advice. XL edited and reviewed the paper.
The authors declare that they have no conflict of interest.
This work was funded by the National Natural Science Foundation of China (grant nos. 31660642 and 31760649), and the Science-technology Support Plan Project of Gansu Province (grant no. 18JR3RA373).
This paper was edited by Steffen Maak and reviewed by three anonymous referees.
Blank, M. F. and Grummt, I.: The seven faces of SIRT7, Transcription, 8, 67–74, https://doi.org/10.1080/21541264.2016.1276658, 2017.
Bosch-Presegué, L. and Vaquero, A.: Sirtuins in stress response: guardians of the genome, Oncogene, 33, 3764–3775, https://doi.org/10.1038/onc.2013.344, 2014.
Cioffi, M., Vallespinos-Serrano, M., Trabulo, S. M., Fernandez-Marcos, P. J., Firment, A. N., Vazquez, B. N., Vieira, C. R., Mulero, F., Camara, J. A., Cronin, U. P., Perez, M., Soriano, J., G. Galvez, B., Castells-Garcia, A., Haage, V., Raj, D., Megias, D., Hahn, S., Serrano, L., Moon, A., Aicher, A., and Heeschen, C.: MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3, Cell Rep., 12, 1594–1605, https://doi.org/10.1016/j.celrep.2015.08.006, 2015.
Cui, Y., Yan, H. L., Wang, K., Xu, H., Zhang, X. L., Zhu, H. J., Liu, J. W., Qu, L., Lan, X. Y., and Pan, C. Y.: Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat, Front. Genet., 9, 91, https://doi.org/10.3389/fgene.2018.00091, 2018.
de Aguiar Vallim, T. Q., Tarling, E. J., Ahn, H., Hagey, L. R., Romanoski, C. E., Lee, R. G., Graham, M. J., Motohashi, H., Yamamoto, M., and Edwards, P. A.: MAFG is a transcriptional repressor of bile acid synthesis and metabolism, Cell Metab., 21, 298–311, https://doi.org/10.1016/j.cmet.2015.01.007, 2015.
Donlon, T. A., Morris, B. J., Chen, R., Masaki, K. H., Allsopp, R. C., Willcox, D. C., Tiirikainen, M., and Willcox, B. J.: Analysis of polymorphisms in 59 potential candidate genes for association with human longevity, J. Gerontol. A. Biol. Sci. Med. Sci., 73, 1459–1464, https://doi.org/10.1093/gerona/glx247, 2018.
Gui, L. S., Xin, X. L., Wang, J. L., Hong, J. Y., and Zan, L. S.: Expression analysis, single nucleotide polymorphisms within SIRT4 and SIRT7 genes and their association with body size and meat quality traits in Qinchuan cattle, J. Integr. Agr., 15, 2819–2826, https://doi.org/10.1016/s2095-3119(16)61419-3, 2016.
Jesús, S., Gómez-Garre, P., Carrillo, F., Cáceres-Redondo, M. T., Huertas-Fernández, I., Bernal-Bernal, I., Bonilla-Toribio, M., Vargas-González, L., Carballo, M., and Mir, P.: Genetic association of sirtuin genes and Parkinson's disease, J. Neurol., 260, 2237–2241, https://doi.org/10.1007/s00415-013-6970-7, 2013.
Jiang, L., Xiong, J., Zhan, J., Yuan, F., Tang, M., Zhang, C., Cao, Z., Chen, Y., Lu, X., Li, Y., Wang, H., Wang, L., Wang, J., Zhu, W. G., and Wang, H.: Ubiquitin-specific peptidase 7 (USP7)-mediated deubiquitination of the histone deacetylase SIRT7 regulates gluconeogenesis, J. Biol. Chem., 292, 13296–13311, https://doi.org/10.1074/jbc.M117.780130, 2017.
Katsuoka, F. and Yamamoto, M.: Small Maf proteins (MafF, MafG, MafK): History, structure and function, Gene, 586, 197–205, https://doi.org/10.1016/j.gene.2016.03.058, 2016.
Komar, A. A.: Genetics. SNPs, silent but not invisible, Science, 315, 466–467, https://doi.org/10.1126/science.1138239, 2007.
Krawczak, M., Reiss, J., and Cooper, D. N.: The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences, Hum. Genet., 90, 41–54, https://doi.org/10.1007/BF00210743, 1992.
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., Lei, C. Z., Zhang, A. L., and Zhang, L. Z.: An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits, Small Ruminant Res., 73, 8–12, https://doi.org/10.1016/j.smallrumres.2006.10.009, 2007.
Li, G., Pan, T., Guo, D., and Li, L. C.: Regulatory variants and disease: the E-cadherin–160C/A SNP as an example, Mol. Biol. Int., 2014, 967565, https://doi.org/10.1155/2014/967565, 2014.
Li, J., Erdenee, S., Zhang, S. L., Wei, Z. Y., Zhang, M., Jin, Y. Y., Wu, H., Chen, H., Sun, X. Z., Xu, H. W., Cai, Y., and Lan, X. Y.: Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep, Prion, 12, 42–53, https://doi.org/10.1080/19336896.2017.1405886, 2018a.
Li, J., Zhang, S. L., Erdenee, S., Sun, X. Z., Dang, R. H., Huang, Y. Z., Lei, C. Z., Chen, H., Xu, H., Cai, Y., and Lan, X. Y.: Nucleotide variants in prion-related protein (testis-specific) gene (PRNT) and effects on Chinese and Mongolian sheep phenotypes, Prion, 12, 185–196, https://doi.org/10.1080/19336896.2018.1467193, 2018b.
Li, X. L. and Kazgan, N.: Mammalian sirtuins and energy metabolism, J. Biol. Sci., 7, 575–587, https://doi.org/10.7150/ijbs.7.575, 2011.
Li, Y., Wu, D. D., Boyko, A. R., Wang, G. D., Wu, S. F., Irwin, D. M., and Zhang, Y. P.: Population variation revealed high-altitude adaptation of Tibetan mastiffs, Mol. Biol. Evol., 31, 1200–1205, https://doi.org/10.1093/molbev/msu070, 2014.
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., He, L., and Shi, Y.: A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn), Cell Res., 19, 519–523, https://doi.org/10.1038/cr.2009.33, 2009.
Liu, G., Zhao, H., Tan, X., Cheng, H., You, W., Wan, F., Liu, Y., Song, E., and Liu, X.: SIRT1 gene polymorphisms associated with carcass traits in Luxi cattle, Arch. Anim. Breed., 60, 27–32, https://doi.org/10.5194/aab-60-27-2017, 2017.
Ma, L., Li, Z. Z., Cai, Y., Xu, H. W., Yang, R. L., and Lan, X. Y.: Genetic variants in fat and short tailed sheep from high-throughput RNA-sequencing data, Anim. Genet., 49, 483–487, https://doi.org/10.1111/age.12699, 2018a.
Ma, L., Zhang, M., Jin, Y. Y., Sarantsetseg, E., Hu, Li. Y., Chen, H., Cai, Y., and Lan, X. Y.: Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep, Front. Genet., 9, 365, https://doi.org/10.3389/fgene.2018.00365, 2018b.
Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C., and Horikawa, I.: Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins, Mol. Biol. Cell., 16, 4623–4635, https://doi.org/10.1091/mbc.e05-01-0033, 2005.
Mu, P., Liu, K, Lin, Q., Yang, W., Liu, D., Lin, Z., Shao, W., and Ji, T.: Sirtuin 7 promotes glioma proliferation and invasion through activation of the ERK/STAT3 signaling pathway, Oncol. Lett., 17, 1445–1452, https://doi.org/10.3892/ol.2018.9800, 2019.
O'Callaghan, C. and Vassilopoulos, A.: Sirtuins at the crossroads of stemness, aging, and cancer, Aging Cell, 16, 1208–1218, https://doi.org/10.1111/acel.12685, 2017.
Roth, M. and Chen, W. Y.: Sorting out functions of sirtuins in cancer, Oncogene, 33, 1609–1620, https://doi.org/10.1038/onc.2013.120, 2014.
Shi, Y. Y. and He, L.: SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci, Cell Res., 15, 97–98, https://doi.org/10.1038/sj.cr.7290272, 2005.
Shin, J. Y. and Chen, D.: Molecular, Cellular, and Physiological Characterization of Sirtuin 7 (SIRT7), Methods Mol. Biol., 1436, 271–277, https://doi.org/10.1007/978-1-4939-3667-0_18, 2016.
Tak, Y. G. and Farnham, P. J.: Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome, Epigenetics Chromatin, 8, 57, https://doi.org/10.1186/s13072-015-0050-4, 2015.
Tang, B. L.: SIRT7 and hepatic lipid metabolism, Front. Cell. Dev. Biol., 3, 1–3, https://doi.org/10.3389/fcell.2015.00001, 2015.
Tang, X., Shi, L., Xie, N., Liu, Z., Qian, M., Meng, F., Xu, Q., Zhou, M., Cao, X., Zhu, W. G., and Liu, B.: SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis, Nat. Commun., 8, 318, https://doi.org/10.1038/s41467-017-00396-9, 2017.
Wang, X. Y., Yang, Q., Wang, K., Zhang, S. H., Pan, C. Y., Chen, H., Qu, L., Yan, H. L., and Lan, X. Y.: A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats, Anim. Genet., 48, 735–736, https://doi.org/10.1111/age.12617, 2017.
Wang, X. Y., Yang, Q., Wang, K., Yan, H. L., Pan, C. Y., Chen, H., Liu, J. W., Zhu, H. J., Qu, L., and Lan, X. Y.: Two strongly linked single nucleotide polymorphisms (Q320P and V397I) in GDF9 gene are associated with litter size in cashmere goats, Theriogenology, 125, 115–121, https://doi.org/10.1016/j.theriogenology.2018.10.013, 2019.
Xu, X. C., Li, B. B., Wei, X., Yang, Y. X., Wang, X. L., and Chen, Y. L.: Differential expression of peroxisome proliferator-activated receptor γ, fatty acid synthase, and hormone-sensitive lipase in fat-tailed and thin-tailed sheep breeds, Genet. Mol. Res., 14, 15624–15633, https://doi.org/10.4238/2015.December.1.14, 2015.
Yamagata, K. and Yoshizawa, T. Transcriptional Regulation of Metabolism by SIRT1 and SIRT7, Int. Rev. Cell Mol. Biol., 335, 143–166, https://doi.org/10.1016/bs.ircmb.2017.07.009, 2018.
Yang, Q., Zhang, S. H., Liu, L. L., Cao, X. K., Lei, C. Z., Qi, X. L., Lin, F. P., Qu, W. D., Qi, X. S., Liu, J. M., Wang, R., Chen, H., and Lan, X. Y.: Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene, Prion, 10, 409–419, https://doi.org/10.1080/19336896.2016.1211593, 2016.
Yang, Q., Yan, H. L., Li, J., Xu, H., Wang, K., Zhu, H. J., Chen, H., Qu, L., and Lan, X. Y.: A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size, Anim. Genet., 48, 499–500, https://doi.org/10.1111/age.12551, 2017.
Ye, X., Li, M. T., Hou, T. Y., Gao, T., Zhu, W. G., and Yang, Y.: Sirtuins in glucose and lipid metabolism, Oncotarget, 8, 1845–1859, https://doi.org/10.18632/oncotarget.12157, 2017.
Yoshizawa, T., Karim, M. F., Sato, Y., Senokuchi, T., Miyata, K., Fukuda, T., Go, C., Tasaki, M., Uchimura, K., Kadomatsu, T., Tian, Z., Smolka, C., Sawa, T., Takeya, M., Tomizawa, K., Ando, Y., Araki, E., Akaike, T., Braun, T., Oike, Y., Bober, E., and Yamagata, K.: SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway, Cell Metab., 19, 712–721, https://doi.org/10.1016/j.cmet.2014.03.006, 2014.
Zhang, S. H., Sun, K., Bian, Y. N., Zhao, Q., Wang, Z., Ji, C. N., and Li, C. T.: Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China, Sci. Rep., 5, 18336, https://doi.org/10.1038/srep18336, 2015.
Zhao, H., He, S., Zhu, Y., Cao, X., Luo, R., Cai, Y., Xu, H., and Sun, X.: A novel 29 bp insertion/deletion (indel) variant of the LHX3 gene and its influence on growth traits in four sheep breeds of various fecundity, Arch. Anim. Breed., 60, 79–85, https://doi.org/10.5194/aab-60-79-2017, 2017.
Zhu, L., Michel, V., and Bakovic, M.: Regulation of the mouse CTP: Phosphoethanolamine cytidylyltransferase gene Pcyt2 during myogenesis, Gene, 447, 51–59, https://doi.org/10.1016/j.gene.2009.07.014, 2009.