the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Copy number variation (CNV) in the IGF1R gene across four cattle breeds and its association with economic traits
Yi-Lei Ma
Yi-Fan Wen
Xiu-Kai Cao
Jie Cheng
Yong-Zhen Huang
Lin-Yong Hu
Chu-Zhao Lei
Xing-Lei Qi
Hui Cao
Hong Chen
The insulin-like growth factor 1 receptor (IGF1R) plays a vital role in immunomodulation and muscle and bone growth. The copy number variation (CNV) is believed to the reason for many complex phenotypic variations. In this paper, we statistically analyzed the copy number and the expression profiling in different tissue types of the IGF1R gene using the 422 samples from four Chinese beef cattle breeds, and the mRNA of IGF1R was widely expressed in nine tissue types of adult cattle (heart, liver, kidney, muscle, fat, stomach, spleen, lung and testis). Results of CNV and growth traits indicated that the IGF1R CNV was significantly associated with body weight and body height of Jinnan (JN) cattle and was significantly associated with body height and hucklebone width of Qinchuan (QC) cattle, making IGF1R CNV a promising molecular marker to improve meat production in beef cattle breeding. Bioinformatics predictions show that the CNV region is highly similar to the human genome, and there are a large number of transcription factors, DNase I hypersensitive sites, and high levels of histone acetylation, suggesting that this region may play a role in transcriptional regulation, providing directions for further study of the role of bovine CNV and economic traits.
- Article
(1472 KB) - Full-text XML
- BibTeX
- EndNote
With the development of industry and the improvement of living standard, beef has gradually made up a greater proportion of the food consumed (Barendse et al., 1997). The economic performance of production traits can be improved by continuous breeding. Various genes regulate meat quality traits, so we can study their genetic variation for marker-assisted selection (MAS) (Cooper et al., 2011). MAS is a breeding method which genotypes molecular markers in larger populations, and these markers are usually linked to causal mutations (Ruane et al., 2007). For example, single nucleotide polymorphism (SNP) and insertion/deletion (Indel) have been used to explore growth traits of cattle in genomic regions (Jin et al., 2016; Pan et al., 2013). However, the above methods also have certain limitations. The effect of many SNPs and Indels, presenting one point or a small mutation, have proved to be insufficient as a cause of phenotype alteration (Huaixing et al., 2008; Sun et al., 2012). So, to speed up molecular breeding, we should choose more effective methods in genomic variations.
Copy number variation (CNV) as a new form of genetic variation is defined as the insertion or deletion of more than 50 bp at the genome level between two individuals of the same species (Mills et al., 2011). In addition, CNV is considered to have influenced many mammalian phenotypes, and it has emerged that the dose effect can affect the expression level of the dose-sensitive genes. This may also be due to other factors such as the fusion of genes, gene function blocking, the location effect, and the removal effect of recessive alleles (Conrad et al., 2010; Kurotaki et al., 2005). CNVs as genomic structural variations contain much more bases, so a stronger genetic impact may arise. In humans, large-scale analyses of CNV data have found that some genes are linked to human diseases, such as C4 in systemic lupus erythematosus, FCGR3B in glomerulonephritis, hBD-1 in psoriasis, and CCL3L1 in HIV/AIDS (Fanciulli and Al, 2007; Gonzalez et al., 2005; Hollox et al., 2008; Yang et al., 2007).
The insulin-like growth factor-1 receptor (IGF1R), a member of IGF gene family, plays essential roles in embryo stages and individual growth after birth (Savage et al., 2010; Ziv and Hu, 2011). IGF1R is the receptor with which IGFs perform biological effects. It can regulate IGF half-life and activity, and it plays a very important role in immune regulation, the formation of lymphocyte, and muscle and bone growth (Adams et al., 2000; Chen et al., 2012). Previous articles have reported that IGF1R gene polymorphism could affect the growth traits of different species, providing theoretical support for genetic improvement (Proskura and Szewczuk, 2014; Roldan et al., 2007; Szewczuk et al., 2013). It is worth mentioning that IGF1R regulates cell proliferation and apoptosis as a vital target to treat cancer (Baserga et al., 2003; Neuzillet et al., 2017). Meanwhile, a high IGF1R gene copy number may have a positive effect on treating cancer (Dziadziuszko et al., 2010).
Thus, we use the IGF1R gene as a candidate gene to study the correlation between the gene copy number variation and the growth traits of Chinese cattle, so as to promote the genetic improvement of beef cattle.
2.1 Samples and trait record
There is currently a commitment to improve the beef quality in Chinese cattle under intensive animal husbandry, and in order to made a thorough inquiry of copy number variation in the bovine IGF1R gene, this study used a total of 422 blood or ear samples of female cattle. The cattle sampled were three endemic beef cattle breeds in China (Qinchuan cattle, QC; Jinnan cattle, JN; Nanyang cattle, NY) and a new beef cattle breed (Xianan cattle, XN) derived from selected crossbreeding between French Charolais (male) and NY cattle (female). The 422 head of cattle were in the same feeding environment and were forage-fed by leisurely grazing from weaning at 6 months of adulthood. None of the cattle were genetically related. Then we used nine kinds of adult QC bovine tissue types (n=3), including heart, liver, kidney, stomach, muscle, lung, spleen and fat for expression pattern analysis. In addition, the body sizes of QC (body height (BH), hip height (HH), body slanting length (BSL), chest width (CW), rump length (RL), hucklebone width (HW), and body weight (BW)), JN (body height (BH), hip height (HH), body slanting length (BSL), chest width (CW), rump length (RL), and body weight (BW)) and XN (body height (BH), hip height (HH), body slanting length (BSL), chest width (CW), cannon circumference (CC), and body weight (BW)) were recorded for further association analysis (Gilbert et al., 1993). All experiments were approved by the Northwest A&F University Ethics Committee.
2.2 Genomic DNA/total RNA isolation
This study used a standard phenol–chloroform protocol to extract genomic DNA from blood and ear tissue (Welter, 1989). As per the instruction manual, total RNA was isolated from tissue by using a TRIzol reagent and treated with RNase-free DNase (TaKaRa, Dalian, China). The quality of RNA was evaluated via 1 % agarose gel electrophoresis, and nanodrop 2000 was used to measure RNA concentration. In addition, this study used PrimeScrip RT Reagent Kit to obtain cDNA (Clontech, TaKaRa). In the end, the DNA and cDNA were diluted to 25 ng µL−1 and then kept in the −80 ∘C freezer.
2.3 Copy number variation and mRNA profiling
Using genomic quantitative polymerase chain reaction (qPCR), the bovine basic transcription factor 3 (BTF3) gene was the housekeeping gene to validate the copy number of IGF1R, and the β-actin gene was used as a housekeeping gene for the tissue distribution profile. In addition, we used the primer 5.0 software to design the primer named IGF1R-P1 from the CNV region of the IGF1R DNA base sequence and to design the primer named IGF1R-P2, which was must span an exon–exon junction from the region of the IGF1R mRNA base sequence. The primer BTF3 and β-actin were also designed by same software (Table 1). It was worth noting that the semiquantitative method was used to measure the quality of primers before qPCR by Bio-Rad CFX 96™ RealTime Detection System (Bio-Rad, Hercules, CA). A total of 12.5 µL reaction mixtures contained 25 ng of genomic DNA/cDNA, 5 pmol of primers 6.25 µL 2*RealStar Green Power Mixture (Genestar, Beijing, China), and 4.25 µL ddH2O. Meanwhile, this system ran at one cycle of 10 min at 95 ∘C and then 40 cycles of 15 s at 95 ∘C, and 1 min s at 60 ∘C. For the melting curve, this was one cycle for 1 min at 95 ∘C and then 1 min at 55 ∘C; then, there was an increase at a rate of 0.5 ∘C per cycle to 95 ∘C. Finally, this was repeated three times for every sample and the mean value of intensity ratios ±SD was calculated for further statistical analysis. Amplification efficiencies for both the target and the internal reference were > 97 % using the standard curve method with four serial dilution points (pooled cDNA or DNA concentration ranging from 500 to 50 pg).
2.4 Bioinformatics prediction of IGF1R CNV
Cellular gene expression was critically determined by DNase I hypersensitive sites and sequence-specific transcription factors (TFs) as well as chromatin modifications. We assumed that these regulatory patterns may be located on the IGF1R CNV locus, so we used the sequence of CNV on Cow June 2014 (Bos_taurus_UMD_3.1.1/bosTau8) assembly compared with the whole-genome sequence on humans February 2009 (GRCh37/hg19) assembly to predicted these regulatory patterns.
2.5 Statistical analysis
This study used the formula to confirm the copy number of IGF1R, where ΔCt was the mean of the target gene minus the mean of the housekeeping gene (Bae et al., 2010). In addition, we used the ANOVA method in the SPSS software to clarify the association between the copy number of the IGF1R gene and the growth traits among the four breeds. According to the hypothesis that there were two copies of DNA in the control region for autosomes, the quantity of the copy number was divided into three types: loss type< 2; normal type: = 2; gain type: > 2 (Yi et al., 2015). Notably, the effects of farm, sex, and season of birth (spring versus fall) did not have any obvious significance for variability of traits in the four breeds as has been previously reported (Cao et al., 2016; Liu et al., 2014, 2016a; Ma et al., 2011). Finally, the following model was used: CNVj+eijk, where Yijk is the observation of the growth traits, μ is the overall mean of each trait, Ai is the effect due to ith age, CNVj is the fixed effect of jth CNV type of IGF1R, and eijk is the random residual error (Xu et al., 2013).
3.1 Distribution of CNVs of IGF1R in four cattle breeds
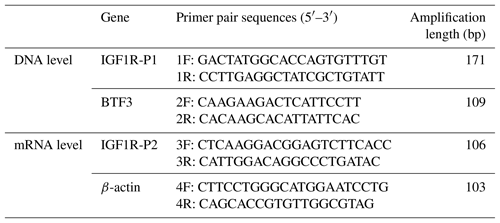
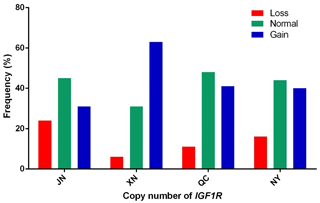
In our previous work, the whole-genome CNV regions were detected by the custom comparative genomic hybridization (CGH) array (NimbleGen, Roche, Madison, WI, USA) in Chinese cattle (Zhang et al., unpublished results). The analysis found that IGF1R covered the CNVR337 region. Specific primers of IGF1R were designed based on the NCBI Bos_taurus_UMD_3.1.1 dbVar assembly. To explore the polymorphism of the copy number (CN) in four cattle breeds, we used two ways to show the condition. The CNV types were classified as loss (CN < 2), normal (CN = 2), and gain (CN > 2) according to . As shown in Fig. 1, the frequencies of copy number polymorphisms in the four cattle breeds had shown which two and three copy numbers take up the large proportion. In addition, as shown in Fig. 2, the three types of IGF1R CNV frequency show two results: the normal type was maximal and the gain type was more frequent than the loss type in JN, QC, and NY breeds all the time, but XN cattle were different from the three breeds in that their gain type was maximal.
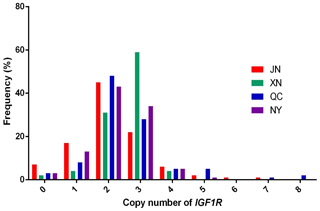
Figure 1Copy number distributions of IGF1R in four Chinese cattle breeds. Histograms show the frequency of individuals with a different copy number of the IGF1R gene. Copy numbers were rounded to the nearest integer.
3.2 Gene expression profiling of IGF1R
IGF1R played an important role in immunomodulation, lymphocyte generation, muscle and bone growth. However, there were few reports of the IGF1R gene expression in cattle, so we used QC tissue to explore gene expression as shown in Fig. 3. We examined nine types of tissue in total in adult cattle: heart, liver, kidney, muscle, fat, stomach, spleen, and lung (from female cattle) and testis (from male cattle). At the adult stage, the mRNA of IGF1R was widely expressed in the nine tissue types, with the highest level of expression in testis. In addition, the level of expression in the kidney, heart, and stomach were second only to the testis, and the middle-level expression was muscle. In the end, low levels of IGF1R were measured in liver, fat spleen, and lung.
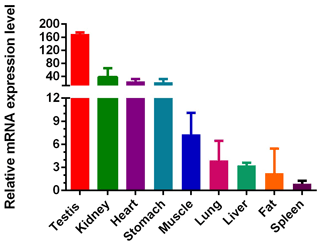
Figure 3Expression profiling of IGF1R in different tissue types of adult cattle. The values are the average of three independent experiments measured by . The relative mRNA expression levels of IGF1R are normalized to β-actin. Testicular tissue is from adult male QC cattle, and other tissue is from adult female QC cattle.
3.3 Correlation analysis of IGF1R CNVs and mRNA expression levels
According to the spatiotemporal expression profiling of the IGF1R gene, we next analyzed the correlation of IGF1R CNVs with mRNA expression levels in testis from 32 adult animals. The reason for using testis was limited by laboratory conditions, and it was highly expressed in male cattle. However, although three types of CNV existed and the mRNA expression varied from 0.17- to 9.43-fold among the tested individuals, the result was found to show no significant correlation between these two data (P=0.079).
3.4 Associations between IGF1R CNVs and growth traits in all cattle
To explore the relationship between the CNV and growth traits, we used genomic DNA from JN (n=128), XN (n=116), QC (n=102), and NY (n=76) cattle. In the analysis, we used more than 100 heads of cattle for analysis in order to provide more compelling evidence and obtain some statistically significant results. In the JN breed, cattle with a copy number loss type had significantly better traits (P<0.05 or P<0.01) than those with normal and gain types, including body height and body weight (Table 2). Similarly, in the QC breed, cattle with a copy number loss type had a significant body height and hucklebone width than those with normal and gain types (P<0.05 or P<0.01) (Table 2). Among XN individuals, no significant association was found at the IGF1R DNA CNV locus with growth traits of 2-year-old cattle (P>0.05) (Table 2), but there was a tendency for loss copy number cases to perform better phenotypic traits than gain or normal copy number cases.
Table 2Association analysis of IGF1R DNA copy number variation (CNV) types with growth traits in cattle.
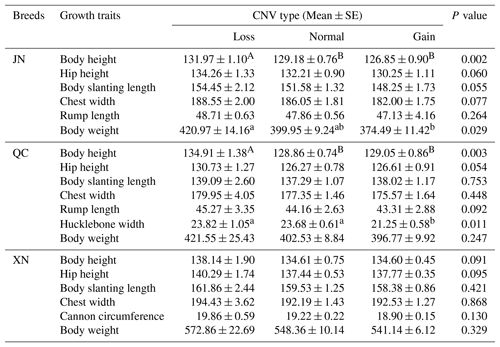
Note: a and b denote values that differ significantly at P<0.05; A and B denote values that differ significantly at P<0.01. JN: loss (31), normal (57), and gain (40). QC: loss (11), normal (49), and gain (42). XN: loss (8), normal (35), and gain (73).
3.5 Bioinformatics prediction of IGF1R CNV
We found that the similarity between the IGF1R CNV sequence in cattle and the whole-genome sequence in humans is 87.5 %, so this region may perform a similar function in the human genome. The result of histone acetylation showed that this region had highly horizontal acetylation. In addition, we found this region had many DNase I hypersensitive sites and transcription factors, and the darker the color, the stronger the ability of transcription factors to bind (Fig. 4).
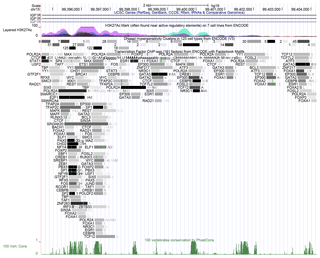
Figure 4Bioinformatics analysis of IGF1R CNV. The peak means histone acetylation above this figure. The rectangle with a number or a letter means DNase I hypersensitive sites or sequence-specific TFs, respectively, and the darker the color, the more likely the combination is. The other peak means similarity between the sequence of IGF1R CNV in cattle and the whole-genome sequence in humans at the bottom of this figure.
With the development of genomics, copy number variation has recently been shown to be associated with disease and phenotypic variation (Bujakowska et al., 2017; Keel et al., 2017; Shi et al., 2016). And it had been continuously confirmed to be able to influence individual phenotypic traits through gene dosage or structural variation (Bickhart et al., 2012; Wright et al., 2009). In cattle, previous research has shown that several CNVs were associated with detoxification, immunizing, and signal recognition; these gene receptors include cytochrome P450, ATP-binding cassette (ABC) transporters, and β-defensins (Hou et al., 2011). In addition, the genes MAPK10, LEPR, and MICAL-L2 within CNVs could influence Chinese cattle's phenotypic variation (Liu et al., 2016; Shi et al., 2015; Xu et al., 2013).Further experiments were conducted to explore the potential role of candidate gene CNV through a large sample of different cattle breeds.
IGF1 is a protein associated with insulin structure and has an important regulatory role in cell growth, differentiation, and maintaining differentiation, whether before or after birth. It is also an important factor affecting weight (Van Laere et al., 2003). Studies have shown that IGF2 plays an important role in the growth and meat quality of pigs (Maagdenberg et al., 2008). IGF1R is a transmembrane glycoprotein with tyrosine kinase activity in both the IGF1 ligands and the IGF2 ligands. Both need to combine IGF1R play antiapoptotic activity, promoting the transformation of cell mitosis, etc. (Pollak, 2008). So, it can be safely assumed that the IGF1R gene may influence bovine phenotype traits. Actually, our results showed that the CNVs of the IGF1R gene were significantly associated with body height, body weight, and hucklebone width in Chinese cattle.
Due to the different genetic backgrounds of the four breeds, a large number of specific CNV loci have been detected in cattle (Liu et al., 2010). Among them, compared with the other three varieties, the XN bovine gain type was more than the other variant types, which may be caused by the difference in breeding background between varieties. XN cattle is a type of beef cattle which is a hybrid of French Charolais and Chinese NY cattle, while the other three varieties of yellow cattle belong to Chinese indigenous local yellow cattle. At the same time, the IGF1R copy number was dispersed in JN cattle, QC cattle, and NY cattle, indicating that the CNV locus had a large degree of variation among these three cattle breeds, which could further influence the phenotype of cattle.
IGF1R and insulin receptor (IR) is homologous proteins with multiple domains of tyrosine kinase. Both receptors are glycoproteins, which are composed of two α and two β subunits. α-Subunits are extracellular and participate in the ligand binding, while β-subunits contain the transmembrane and intracellular domain (Keyhanfar et al., 2007; Lawrence et al., 2007). IGF1R binds IGF1 with high affinity, while it has a low affinity for IGF2 and insulin (INS). Activated IGF1R is involved in cell growth and survival control. IGF1R is essential for tumor transformation and the survival of malignant cells. Ligand binding activated receptor kinase, which leads to receptor phosphorylation and a variety of substrates of tyrosine phosphorylation as signal transduction proteins including insulin receptor substrate (IRS1/2), Shc, and 14-3-3 proteins. Phosphorylation of IRS protein leads to the activation of two major signaling pathways: the PI3K-AKT/PKB pathway and the Ras-MAPK pathway. The result of activating the MAPK pathway is increased cell proliferation, while activating the PI3K pathway inhibits cell apoptosis and stimulates protein synthesis (Gkioka et al., 2015). So far, there has been little information on IGF1/IGF1R signaling, especially in cattle. In this study, the cattle IGF1R gene also showed a broad spectrum in expression, indicating that IGF1R is involved in the transmission and regulation of the growth axis in multiple tissue types. So, it is likely to be involved in a variety of regulation, leading to a change in adult individual metabolic activity and fat deposition ultimately reflected on the phenotype of the cattle. In addition, we think the CNV locus can influence Chinese cattle although it is located in intron 1. With the development of studies, it has been recognized that through the alternative splicing of introns, a single gene can encode many different proteins at the same time. Recently, it has become increasingly clear that introns and their shear processes can influence gene expression through gene transcription, mRNA transport, localization, and translation (Bicknell et al., 2012; Hir et al., 2003).
Dosage effect is one important mechanism underlying the phenotypic effects of CNV. A corresponding missing or amplification may cause function disorder, which lacks a corresponding cause to reduce the level of gene expression. Amplification results in an increase in expression level. Previous studies found that the oncogenes Myc and MYCN were at 17q21.31, chr8q24.22 ∼ 24.23, and significant DNA fragment amplification was observed in this region. Deficiency was observed at 9p21.1 ∼ 9p21.3 and 6p23.1 (Lu et al., 2009). In addition, the dose effect of CNV can also affect the penetrance of genes (Beckmann et al., 2007). Although the correlation analysis was carried out, there is no significant correlation between IGF1R CNVs and mRNA expression levels. CNV may influence bovine growth traits by other regulatory mechanisms. Previous studies indicate that CNV may be a key factor in reducing the penetrance of some pathogenic genes and can also significantly influence the expression products of genes by changing their structure. A study found that CNV was associated with the susceptibility of neuroblastoma at lq21.1, which led to changes in the expression of NBPF transcription NBPF23 (Diskin et al., 2009). Meanwhile, bioinformatics predicted that there are a lot of transcription factors in the region, such as FOSL2, SP1, MYC, and E2F1. They play a key role in growth and development (Schwarz et al., 1995; Black et al., 1999; Fan et al., 2014). In addition, there are a lot of DNase I hypersensitive sites and high levels of acetylation in this region, which presumably indicates the presence of enhancer regulation in this region. Moreover, it was found that CNV was not significantly correlated with gene expression, indicating that the copy number variation achieved its phenotypic effects through genes other than IGF1R. Further study will be undertaken to outline the underpinning mechanism.
In conclusion, we determined the IGF1R CNV in Chinese cattle breeds. Our study provided a preliminary result for the functional role of the IGF1R CNVs in larger populations and different cattle breeds and for a novel and important marker in cattle breeding programs.
The original data are available upon request to the corresponding author.
YLM and YFW contributed equally to this work.
The authors declare that they have no conflict of interest.
This work was supported by the National Natural Science Foundation of China (no. 31772574, 31601926), the Program of National Beef Cattle and Yak Industrial Technology System [CARS-37], Special Fund of Xinyang Normal University (no. 2017001) Science and Technology Plan Project of Yangling Demonstration Area in 2018, Applied Basic Research Program of Qinghai Province(2014-ZJ-710).
This paper was edited by Steffen Maak and reviewed by Runfeng Zhang and one anonymous referee.
Adams, T. E., Epa, V. C., Garrett, T. P. J., and Ward, C. W.: Structure and function of the type 1 insulin-like growth factor receptor, Cell. Mol. Life Sci., 57, 1050, https://doi.org/10.1007/PL00000744, 2000.
Bae, J. S., Cheong, H. S., Kim, L. H., Namgung, S., Park, T. J., Chun, J. Y., Kim, J. Y., Pasaje, C. F., Lee, J. S., and Shin, H. D.: Identification of copy number variations and common deletion polymorphisms in cattle, Bmc Genomics, 11, 232, https://doi.org/10.1186/1471-2164-11-232, 2010.
Barendse, W., Vaiman, D., Kemp, S. J., Sugimoto, Y., Armitage, S. M., Williams, J. L., Sun, H. S., Eggen, A., Agaba, M., Aleyasin, S. A., Band, M., Bishop, M. D., Buitkamp, J., Byrne, K., Collins, F., Cooper, L., Coppettiers, W., Denys, B., Drinkwater, R. D., Easterday, K., Elduque, C., Ennis, S., Erhardt, G., Li, L., and Et, A.: A medium-density genetic linkage map of the bovine genome, Mamm. Genome, 8, 21–28, 1997.
Baserga, R., Peruzzi, F., and Reiss, K.: The IGF-1 receptor in cancer biology, Int. J. Cancer, 107, 873–877, 2003.
Beckmann, J. S., Estivill, X., and Antonarakis, S. E.: Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability, Nat. Rev. Genet. 8, 639–646, 2007.
Bickhart, D. M., Hou, Y., Schroeder, S. G., Alkan, C., Cardone, M. F., Matukumalli, L. K., Song, J., Schnabel, R. D., Ventura, M., and Taylor, J. F.: Copy number variation of individual cattle genomes using next-generation sequencing, Genome Res., 22, 778–790, 2012.
Bicknell, A. A., Cenik, C., Chua, H. N., Roth, F. P., and Moore, M. J.: Introns in UTRs: Why we should stop ignoring them, Bioessays News & Reviews in Molecular Cellular & Developmental Biology, 34, 1025–1034, 2012.
Black, A. R., Jensen, D., Lin, S. Y., and Azizkhan, J. C.: Growth/cell cycle regulation of Sp1 phosphorylation, J. Biol. Chem., 274, 1207, https://doi.org/10.1074/jbc.274.3.1207, 1999.
Bujakowska, K. M., Fernandez-Godino, R., Place, E., Consugar, M., Navarro-Gomez, D., White, J., Bedoukian, E. C., Zhu, X., Xie, H. M., and Gai, X.: Copy-number variation is an important contributor to the genetic causality of inherited retinal degenerations, Genetics in Medicine Official Journal of the American College of Medical Genetics, 19, 643–651, 2017.
Cao, X. K., Zhan, Z. Y., Huang, Y. Z., Lan, X. Y., Lei, C. Z., Qi, X. L., and Chen, H.: Variants and haplotypes within MEF2C gene influence stature of chinese native cattle including body dimensions and weight, Livest. Sci., 185, 106–109, 2016.
Chen, W., Feng, Y., Zhao, Q., Zhu, Z., and Dimitrov, D. S.: Human monoclonal antibodies targeting nonoverlapping epitopes on insulin-like growth factor II as a novel type of candidate cancer therapeutics, Mol. Cancer Ther., 11, 1400–1410, 2012.
Conrad, D. F., Pinto, D., Redon, R., Feuk, L., Gokcumen, O., Zhang, Y., Aerts, J., Andrews, T. D., Barnes, C., and Campbell, P.: Origins and functional impact of copy number variation in the human genome, Nature, 464, 704–712, 2010.
Cooper, G. M., Coe, B. P., Girirajan, S., Rosenfeld, J. A., Vu, T. H., Baker, C., Williams, C., Stalker, H., Hamid, R., Hannig, V., Abdel-Hamid, H., Bader, P., McCracken, E., Niyazov, D., Leppig, K., Thiese, H., Hummel, M., Alexander, N., Gorski, J., Kussmann, J., Shashi, V., Johnson, K., Rehder, C., Ballif, B. C., Shaffer, L. G., and Eichler, E. E.: A copy number variation morbidity map of developmental delay, Nat. Genet., 43, 838–846, 2011.
Diskin, S. J., Hou, C., Glessner, J. T., Attiyeh, E. F., Laudenslager, M., Bosse, K., Cole, K., Mossé, Y. P., Wood, A., and Lynch, J. E.: Copy number variation at 1q21.1 associated with neuroblastoma, Nature, 459, 987–991, 2009.
Dziadziuszko, R., Merrick, D. T., Witta, S. E., Mendoza, A. D., Szostakiewicz, B., Szymanowska, A., Rzyman, W., Dziadziuszko, K., Jassem, J., and Jr, B. P.: Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression, Journal of Clinical Oncology Official, Journal of the American Society of Clinical Oncology, 28, 2174, https://doi.org/10.1200/jco.2009.24.6611, 2010.
Fanciulli, M. and Al, E.: FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity, Nat. Genet., 39, 721–723, 2007.
Gilbert, R. P., Bailey, D. R., and Shannon, N. H.: Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets, J. Anim. Sci., 71, 1712, https://doi.org/10.2527/1993.7171712x, 1993.
Gkioka, E., Msaouel, P., Philippou, A., Vlaghogiannis, N. I., Vogkou, C. T., Margiolis, A., and Koutsilieris, M.: Review: The Role of Insulin-like Growth Factor-1 Signaling Pathways in Uterine Leiomyoma, Vivo, 29, 637–650, 2015.
Gonzalez, E., Kulkarni, H., Bolivar, H., Mangano, A., Sanchez, R., Catano, G., Nibbs, R. J., Freedman, B. I., Quinones, M. P., and Bamshad, M. J.: The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility, Science, 307, 1434, https://doi.org/10.1126/science.1101160, 2005.
Huaixing, L., Ying, W., Loos, R. J. F., Hu, F. B., Yong, L., Jing, W., Zhijie, Y., and Xu, L.: Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population, Diabetes, 57, 264–268, 2008.
Hir, H. L., Nott, A., and Moore, M. J.: How introns influence and enhance eukaryotic gene expression?, Trends Biochem. Sci., 28, 215–220, 2003.
Hollox, E. J., Huffmeier, U., Zeeuwen, P. L. J. M., Palla, R., Lascorz, J., Rodijkolthuis, D., Kerkhof, P. C. M. V., Traupe, H., Jongh, G. D., and Heijer, M. D.: Psoriasis is associated with increased beta-defensin genomic copy number, Nat. Genet., 40, 23–25, 2008.
Hou, Y., Liu, G. E., Bickhart, D. M., Cardone, M. F., Kai, W., Kim, E. S., Matukumalli, L. K., Ventura, M., Song, J., and Vanraden, P. M.: Genomic characteristics of cattle copy number variations, Bmc Genomics, 12, 127, https://doi.org/10.1186/1471-2164-12-127, 2011.
Jin, Y., Cai, H., Liu, J., Lin, F., Qi, X., Bai, Y., Lei, C., Chen, H., and Lan, X.: The 10 bp duplication insertion/deletion in the promoter region within paired box 7 gene is associated with growth traits in cattle, Arch. Anim. Breed., 59, 469–476, https://doi.org/10.5194/aab-59-469-2016, 2016.
Keel, B. N., Keele, J. W., and Snelling, W. M.: Genome-wide copy number variation in the bovine genome detected using low coverage sequence of popular beef breeds, Anim. Genet., 48, 141–150, 2017.
Keyhanfar, M., Booker, G. W., Whittaker, J., Wallace, J. C., and Forbes, B. E.: Precise mapping of an IGF-I-binding site on the IGF-1R, Biochem. J., 401, 269–277, 2007.
Kurotaki, N., Shen, J. J., Touyama, M., Kondoh, T., Visser, R., Ozaki, T., Nishimoto, J., Shiihara, T., Uetake, K., and Makita, Y.: Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency, Genetics in Medicine Official Journal of the American College of Medical Genetics, 7, 479–483, 2005.
Lawrence, M. C., Mckern, N. M., and Ward, C. W.: Insulin receptor structure and its implications for the IGF-1 receptor, Curr. Opin. Struct. Biol., 17, 699–705, 2007.
Liu, G. E., Hou, Y., Zhu, B., Cardone, M. F., Jiang, L., Cellamare, A., Mitra, A., Alexander, L. J., Coutinho, L. L., and Dell'Aquila, M. E.: Analysis of copy number variations among diverse cattle breeds, Genome Res., 20, 693–703, 2010.
Liu, M., Li, B., Huang, Y., Yang, M., Lan, X., Lei, C., Qu, W., Bai, Y., and Chen, H.: Copy number variation of bovine MAPK10 modulates the transcriptional activity and affects growth traits, Livest. Sci., 194, 44–50, 2016a.
Liu, M., Li, M., Wang, S., Xu, Y., Lan, X., Li, Z., Lei, C., Yang, D., Jia, Y., and Chen, H.: Association analysis of bovine Foxa2 gene single sequence variant and haplotype combinations with growth traits in Chinese cattle, Gene, 536, 385–392, 2014.
Liu, M., Liu, M., Li, B., Zhou, Y., Huang, Y., Lan, X., Qu, W., Qi, X., Bai, Y., and Chen, H.: Polymorphisms of FLII implicate gene expressions and growth traits in Chinese cattle, Molecular & Cellular Probes, 30, 266–272, 2016.
Lu, Y., Ryan, S. L., Elliott, D. J., Bignell, G. R., Futreal, P. A., Ellison, D. W., Bailey, S., and Clifford, S. C.: Amplification and Overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in Medulloblastoma, Plos One, 4, e6159, https://doi.org/10.1371/journal.pone.0006159, 2009.
Ma, L., Qu, Y. J., Huai, Y. T., Li, Z. J., Wang, J., Lan, X. Y., Zhang, C. L., Wang, J. Q., and Chen, H.: Polymorphisms identification and associations of KLF7 gene with cattle growth traits, Livest. Sci., 135, 1–7, 2011.
Maagdenberg, K. V. D., Stinckens, A., Claeys, E., Buys, N., and Smet, S. D.: Effect of the insulin-like growth factor-II and RYR1 genotype in pigs on carcass and meat quality traits, Meat Sci., 80, 293–303, 2008.
Mills, R. E., Walter, K., Stewart, C., Handsaker, R. E., Chen, K., Alkan, C., Abyzov, A., Yoon, S. C., Ye, K., and Cheetham, R. K.: Mapping copy number variation by population-scale genome sequencing, Nature, 470, 59–65, 2011.
Neuzillet, Y., Chapeaublanc, E., Krucker, C., De, K. L., Lebret, T., Radvanyi, F., and Bernard-Pierrot, I.: IGF1R activation and the in vitro antiproliferative efficacy of IGF1R inhibitor are inversely correlated with IGFBP5 expression in bladder cancer, Bmc Cancer, 17, 636, https://doi.org/10.1186/s12885-017-3618-5, 2017.
Pan, C., Wu, C., Jia, W., Xu, Y., Lei, C., Hu, S., Lan, X., and Chen, H.: A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle, Gene, 531, 398–402, 2013.
Pollak, M.: Targeting insulin and insulin-like growth factor signalling in oncology, Curr. Opin. Pharmacol., 8, 384–392, 2008.
Proskura, W. S. and Szewczuk, M.: The polymorphism in the IGF1R gene is associated with body weight and average daily weight gain in Pomeranian Coarsewool ewes, Pak. Vet. J., 34, 514–517, 2014.
Roldan, M. B., White, C., and Witchel, S. F.: Association of the GAA1013-GAG polymorphism of the insulin-like growth factor-1 receptor (IGF1R) gene with premature pubarche, Fertil. Steril., 88, 410–417, 2007.
Ruane, J., Sonnino, A., Guimarães, E. P., Ruane, J., Scherf, B. D., Sonnino, A., and Dargie, J. D.: Marker-assisted selection as a tool for genetic improvement of crops, livestock, forestry and fish in developing countries: an overview of the issues, http://pdfs.semanticscholar.org/56d2/07abda8bedc8e8ec97a6af3366ca5cfe52cb.pdf, 2007.
Savage, M. O., Burren, C. P., and Rosenfeld, R. G.: The continuum of growth hormone-IGF-I axis defects causing short stature: diagnostic and therapeutic challenges, Clin. Endocrinol., 72, 721–728, 2010.
Schwarz, J. K., Bassing, C. H., Kovesdi, I., Datto, M. B., Blazing, M., George, S., Wang, X. F., and Nevins, J. R.: Expression of the E2F1 transcription factor overcomes type beta transforming growth factor-mediated growth suppression, P. Natl. Acad. Sci. USA, 92, 483–487, 1995.
Shi, T., Xu, Y., Yang, M., Huang, Y., Lan, X., Lei, C., Qi, X., Yang, X., and Chen, H.: Copy number variations at LEPR gene locus associated with gene expression and phenotypic traits in Chinese cattle, Anim. Sci. J., 87, 336–343, 2016.
Sun, J., Xue, J., Zhang, C., Lan, X., Lei, C., and Chen, H.: Haplotype combination of the caprine PC1 gene sequence variants and association with growth traits in Chinese Haimen breed, J. Genet., 91, 1–6, 2012.
Szewczuk, M., Zych, S., Wójcik, J., and Czerniawska-Piątkowska, E.: Association of two SNPs in the coding region of the insulin-like growth factor 1 receptor (IGF1R) gene with growth-related traits in Angus cattle, J. Appl. Genet., 54, 305–308, 2013.
Van Laere, A. S., Nguyen, M., Braunschweig, M., Nezer, C., Collette, C., Moreau, L., Archibald, A. L., Haley, C. S., Buys, N., Tally, M., Andersson, G., Georges, M., and Andersson, L.: A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig, Nature, 425, 832–836, https://doi.org/10.1038/nature02064, 2003.
Welter, C.: An efficient salt-chloroform extraction of DNA from blood and tissues, Trends Genet., 5, 389–391, 1989.
Wright, D., Boije, H., Meadows, J. R. S., Bed'Hom, B., Gourichon, D., Vieaud, A., Tixierboichard, M., Rubin, C. J., Imsland, F., and Hallböök, F.: Copy number variation in intron 1 of SOX5 causes the pea-comb phenotype in chickens, Plos Genet., 5, e1000512, https://doi.org/10.1016/0168-9525(89)90181-9, 2009
Xu, Y., Zhang, L., Shi, T., Zhou, Y., Cai, H., Lan, X., Zhang, C., Lei, C., and Chen, H.: Copy number variations of MICAL-L2 shaping gene expression contribute to different phenotypes of cattle, Mamm. Genome, 24, 508–516, 2013.
Yang, Y., Chung, E. K., Wu, Y. L., Savelli, S. L., Nagaraja, H. N., Zhou, B., Hebert, M., Jones, K. N., Shu, Y., and Kitzmiller, K.: Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European American, Am. J. Hum. Genet., 80, 1037, https://doi.org/10.1086/518257, 2007.
Yi, G., Qu, L., Chen, S., Xu, G., and Yang, N.: Genome-wide copy number profiling using high-density SNP array in chickens, Anim. Genet., 46, 148–157, 2015.
Ziv, E. and Hu, D.: Genetic variation in insulin/IGF-1 signaling pathways and longevity?, Ageing Res. Rev., 10, 201–204, 2011.