the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Effects of polymorphisms in CAPN1 and CAST genes on meat tenderness of Chinese Simmental cattle
Xiaomei Sun
Xiuxiang Wu
Yongliang Fan
Yongjiang Mao
Dejun Ji
Bizhi Huang
Zhangping Yang
Considerable evidence has demonstrated that the μ-calpain (CAPN1) gene and its inhibitor calpastatin (CAST) gene are major factors affecting meat quality. Marker-assisted selection (MAS) has been widely used to improve beef quality traits. Therefore, the objective of the present study was to investigate the single nucleotide polymorphisms (SNPs) of bovine CAPN1 and CAST genes using 367 animals representing the four main Chinese cattle breeds and to explore the effects of these SNPs on meat quality traits. Two SNPs within CAPN1 and one SNP in CAST were successfully identified in cattle. Genetic diversity analyses suggested that most SNPs in the four breeds exhibited a moderate genetic diversity. Moreover, associations between individual markers and meat quality traits were analyzed in Chinese Simmental cattle. The CAPN1 4558 A > G locus was found to be significantly associated with shear force value (SFV) and marbling score (BMS), and CAPN1 4684 C > T exerted a significant effect on SFV, while the CAST genotype was not significantly associated with any of the measured traits. SFV, commonly used to measure meat tenderness, represents an important quality trait as it contributes to the flavor of cooked meat. This work confirms the effect of CAPN1 on beef tenderness and lays an important foundation for future cattle breeding.
- Article
(1139 KB) - Full-text XML
-
Supplement
(219 KB) - BibTeX
- EndNote
Marbling and meat color are important meat quality traits that are usually used as an indicator of freshness and hygienic beef products (Castro et al., 2016). In addition, tenderness is considered as the most momentous parameter regarding eating quality (Moeller et al., 2010; Koohmaraie and Geesink, 2006; Kamruzzaman et al., 2013). Thus, color, marbling, tenderness, and flavor contribute importantly to various aspects of beef quality. Currently, consumer demand for high-quality meat is increasing, but the breeding and production of high-quality beef cattle cannot keep pace with it. Meat quality traits are normally influenced by the interaction of many factors, including the animal genetics. Single nucleotide polymorphism markers that can be measured in one or more populations play a central role in modern genetic breeding (Davey et al., 2011; Coll et al., 2014; Melo et al., 2017; Renaville et al., 2015) and are important tools for beef cattle breeding (Li et al., 2013a; Sun et al., 2013; Buzanskas et al., 2017). Characterization of the genes affecting important economical traits and the availability of the bovine genome sequence have allowed researchers to identify the polymorphisms associated with phenotype.
In cattle, genetic markers affecting meat color and beef marbling have rarely been studied, suggesting that the research is urgently needed. Several candidate genes have now been nominated with significant effects on meat tenderness. These include the protease μ-calpain (CAPN1) and its inhibitor calpastatin (CAST) (Barendse et al., 2007; Tait et al., 2014; Gandolfi et al., 2011; Schenkel et al., 2006). The CAPN1 gene, encoding the larger subunit of the cysteine related to the postmortem tenderization process, was located on BTA29, which contains quantitative trait loci of meat quality. CAST, the inhibitor calpastatin of CAPN1 (Koohmaraie, 1996), was also identified as a major factor affecting postmortem tenderization in meat (Enriquez-Valencia et al., 2017). Genetic marker tests for these proteins have been identified for meat tenderness and are now commercially available (Johnston and Graser, 2010). Some genetic markers with positive effects on certain traits are allowed to be incorporated into multi-trait breeding projects.
Discovery of the DNA markers associated with meat quality traits is becoming an interest in cattle breeding. Hence, in this study, CAPN1 and CAST were selected as candidate genes to estimate the frequency of single nucleotide polymorphisms (SNPs) in four Chinese cattle breeds and to determine if individual SNPs were associated with meat quality traits in Chinese Simmental cattle. The results provide potential useful genetic information for beef quality trait improvements.
2.1 Samples and meat quality traits
Genomic DNA samples (n=367) were collected from four Chinese cattle breeds: Chinese Simmental cattle (Sim, n=132), Leiqiong cattle (LQ, n=74), Yunnan high pump (YNH, n=80), and BMY cattle (1∕2 Brahman, 1∕4 Murray Grey, and 1∕4 Yunnan Yellow cattle, n=81). All experiments performed in this study were approved by the Yangzhou University Animal Care and Use Committee (SYXK(SU)2017-0044). The care and use of experimental animals fully complied with local animal welfare laws, guidelines, and policies. Detailed information about the four populations is presented in Table 1. Four cattle breeds were used to investigate the genetic diversity of CAPN1 and CAST genes. The Chinese Simmental cattle, a cross-bred beef cattle, was selected for evaluating the effect of CAPN1 and CAST polymorphisms on meat quality traits and 132 muscle samples were collected from the Chinese Simmental cattle. Four meat quality traits, including shear force value (SFV), marbling score (BMS), meat color score (MCS), and fat color score (FCS), were recorded form 132 Chinese Simmental individuals.
Warner–Bratzler shear force values (SFVs) is one of the standard tools to quantify meat tenderness. SFV was measured on the interface of the longissimus muscle from the 12th and 13th ribs after 24–48 h of aging. The muscle samples were sliced into 2.5 cm thickness parallel to the longitudinal orientation of the muscle fibers and heated in a water bath until the center temperature reached 75 ∘C. After chilling, six cores should be obtained from each sample. Round cores should be 1.27 cm in diameter and removed parallel to the longitudinal orientation of the muscle fibers so that the shearing action is perpendicular to the longitudinal orientation of the muscle fibers. Samples that are not uniform in diameter or have obvious connective tissue defects would not be representative of the sample and should be discarded. Muscle samples that meet the requirement were then measured for shear force values immediately using an XL1155 shear force detector (Xielikeji Co., Ltd, Harbin, China). Beef marbling score (BMS) was evaluated at the longissimus muscle of the 12th and 13th ribs using photographic standards.
2.2 SNP identification and genotyping
Genomic DNA was extracted from the blood samples following the standard procedures (Sambrook and Russell, 2001). The quantity and quality of DNA were measured by a spectrophotometer at 260∕280 nm using an Eppendorf BioPhotometer. Each genome DNA sample was diluted to 50 ng µL−1. The PCR–single strand conformation polymorphism (PCR-SSCP) method was applied to detect the SNPs of two genes. Primers used for SNP identification and genotyping were synthesized by Shanghai Sangon (Table S1 in the Supplement). PCR amplification was performed using a GeneAmp PCR system 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA), with the following protocol: initial denaturation at 94∘ for 5 min, followed by 30 cycles at 94∘ for 30 s, 65∘ for 30 s, and 72∘ for 40 s, with a final extension at 72∘ for 7 min.
A total of 2.0 µL PCR product was mixed with 8 µL of the denaturation solution (50 mmol L−1 NaOH, 1 mmol L−1 EDTA) and 1 µL of the loading buffer containing 0.25 % bromophenol blue and 0.25 % xylene cyanol, denatured for 10 min at 98∘, and rapidly chilled to −20∘. The samples were then detected in 12 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A thermostatically controlled refrigerated circulator was used to maintain constant temperature (4∘) of the gels. The gels were run in the following conditions: preelectrophoresis under 250 V and 40 mA for 10 min; and 150 V and 24 mA for 8 h. The gels were then stained by Silver Stain (Kucharczyk Techniki Elektroforetyczne). The patterns of DNA bands were observed and photographed with the GDS7500 system (UVP). Three samples of each genotype were selected for sequencing with the ABI-PRISM3730 analyzer.
2.3 Statistical analysis
Allele and genotype frequencies were calculated using PopGene (Ver. 3.2) and deviations from Hardy–Weinberg equilibrium (HWE) were checked by chi-square test. The population genetic indexes including gene heterozygosity (He), effective allele numbers (Ne), and polymorphism information content (PIC) were calculated by Nei's method (Nei and Roychoudhury, 1974).
The data were analyzed with the SPSS software with the following fixed-effects model. Multivariate analysis of variance was applied to study the effect of genotypes on meat quality traits within Sim cattle (n=132) using the general linear model (GLM), with genotype serving as fixed effects and BMS, SFV, MCS, and FCS as dependent variables:
where y represents the phenotype records of meat quality traits (e.g., the IMF, BMS, SFV, MCS, and FCS), μ is general mean, Gj is the fixed effect of the genotype (CAPN1 4558, CAPN1 4684, and CAST 596), and e is the random error. The statistics were presented as probability values and least square means ± standard error, and P values < 0.05 were considered statistically significant.
3.1 SNP identification and genotyping
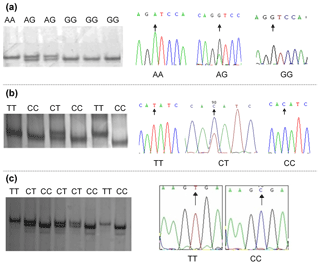
Polymorphisms were detected by DNA pool sequencing. Investigation of exon 1 to exon 6 of the bovine CAPN1 gene and exon 9 of the CAST gene identified three SNPs, and they were located in exon 2 and exon 4 of CAPN1 and exon 9 of CAST, respectively. These SNPs, including CAPN1 4558 A > G, CAPN1 4684 C > T, and CAST 596 T > C, were successfully genotyped by PCR-SSCP (Fig. 1a, b, and c). All SNPs contained two alleles and three genotypes in each population.
3.2 Genetic diversity analyses among four Chinese cattle populations
Genetic diversity analyses are given in Table 2. Allele frequencies, genotype frequencies, and PIC at all SNPs were similar in LQ and YNP, two Bos indicus cattle breeds.
At the CAPN1 4558 A > G locus, the A allele was predominant in all populations, with frequencies ranging from 0.664 to 0.889 within the four beef breeds (Sim, LQ, YNP, and BMY). At the CAPN1 4684 C > T locus, the C allele was predominant in Sim, LQ, and YNP with frequencies varying from 0.516 to 0.818, while the frequency of the C allele in BMY was 0.470. At the CAST 596 T > C locus, the T allele was predominant in all populations, with frequencies ranging from 0.529 to 0.626 within the four beef breeds.
PIC analysis suggested that at the CAPN1 4684 C > T and CAST 596 T > C loci, all four breeds had a moderate genetic diversity (0.250 < PIC < 0.500). At CAPN1 4558 A > G loci, the Sim breed had a moderate genetic diversity, while LQ, YNP, and BMY had a low genetic diversity (PIC < 0.250). Moreover, the χ2 test showed that the genotype distributions of all SNP loci within Chinese Simmental cattle breed and most SNP loci in the other three breeds were in HWE (P>0.05, Table 2). This indicated that the four Chinese cattle populations, especially the Chinese Simmental cattle breed, have experienced low selection pressure and therefore may have great genetic potential in cattle breeding.
Table 2Genotype, allele frequencies, and genetic diversity parameters of bovine CAPN1 and CAST in four Chinese cattle populations.
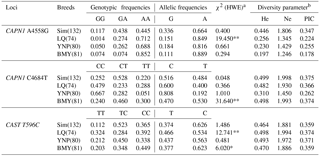
Note: , ; * means P<0.05, ** P<0.01. a χ2 (HWE), Hardy–Weinberg equilibrium χ2 value. b He, gene heterozygosity; Ne, effective allele numbers; PIC, polymorphism information content.
3.3 Association of genotype with meat quality traits in Chinese Simmental cattle
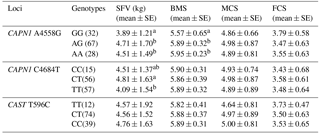
The relationships between the CAPN1 and CAST genotypes and meat quality traits were analyzed in Chinese Simmental cattle (n=132). Traits evaluated were shear force value (SFV), marbling score (BMS), meat color score (MCS), and fat color score (FCS). The results are exhibited in Table 3. At the CAPN1 4558 A > G locus, cattle with the GG genotype displayed lower shear force value than individuals with AG and AA genotypes (P<0.05). At the CAPN1 4684 C > T loci, animals with the TT genotype had significantly lower shear force value than those with the CC and CT genotype (P<0.05). In addition, the CAPN1 4684 C > T locus was found to be significantly association with BMS (P < 0.05), while the CAST 596 T > C loci had no significant association with any of the measured traits (P>0.05).
Consumers are increasingly aware of dietary nutrition, and beef is considered to be a highly nutritious food that contains biological value protein and micronutrients (Scollan et al., 2014). Thus, these trends are also occurring in the fresh beef market as reflected by the demand for higher-quality meat. Meat quality is difficult to define because it is a complex concept affected by various intrinsic qualities (for example, color, tenderness, juiciness, flavor, chemical composition) and extrinsic qualities (brand, quality mark, production environment) (Joo et al., 2013). Knowledge of the above factors has augmented consumer interest in food quality. Beef tenderness is a critical component of palatability, but the difficulty in obtaining phenotypic data until slaughter has made it hard to select for this trait. Therefore, marker-assisted selection (MAS) for genetic improvement could bypass this obstacle if appropriate markers could be found. Thus, increasing attention has been given to improving beef quality traits through MAS.
Considerable evidence has demonstrated that allelic variation in the CAPN1 gene is important in the successful application of MAS in beef cattle populations (Schenkel et al., 2006). It plays an important role in meat tenderness and indirectly acts on other meat quality traits. In the same process, CAST appears to be an inhibiting enzyme of CAPN1. In beef cattle breeding, genetic variation of meat tenderness, structure, and color plays important roles in the MAS (Lee et al., 2014). Therefore, acquiring a highly efficient system for identifying the molecular markers of these genes would constitute an important support in the selection of cattle to produce better beef.
In the present study, SNPs in CAPN1 and CAST were identified and analyzed for associations with meat quality traits in Chinese Simmental cattle. Briefly, the presence of CAPN1 markers had significant effects on beef shear force but no detectable effects were found for the CAST SNP, which agrees with a previous study on beef cattle (Corva et al., 2007). Tenderness, measured by shear force, represents a vital beef quality trait as it contributes to the flavor of cooked meat (Kamruzzaman et al., 2013). Several research studies have been applied to the effects of genetic markers in CAPN1 on beef tenderness (Allais et al., 2011; Curi et al., 2009). In addition, previous studies showed that SNPs in the CAPN1 gene were associated with the level of beef marbling (Li et al., 2013b). Our data illustrated that CAPN1 4558 A > G was found to be significantly associated with BMS and SFV, and CAPN1 4684 C > T was found to exert a significant effect on SFV in Chinese Simmental cattle (* p <0.05). Hence, these genetic markers may be useful in selecting populations for beef breeding.
As a representative of the calpain system, CAST was recognized as a candidate marker in beef breeding with MAS. Herein, no significant association between identified SNPs of the CAST gene and meat quality traits was found in the Chinese Simmental cattle population. This result was consistent with previous research performed on beef cattle from Argentina and on sheep (Corva et al., 2007; Zhou et al., 2008), but disagreed with some research showing that the CAST genotype was associated with meat tenderness in other cattle populations (Casas et al., 2006; Allais et al., 2011; Calvo et al., 2014; Enriquez-Valencia et al., 2017), indicating that this SNP may not be a powerful marker for meat quality traits in Chinese Simmental cattle.
In conclusion, this study supports previous work that CAPN1 genetic markers have a positive effect on meat tenderness and BMS traits, the most important meat quality traits for consumers. The associations between the genotyped SNPs in this research and meat quality traits are of potential interest to the beef industry and could lay an important foundation for the expanding panel of functional variation relevant to meat quality.
Data are available upon reasonable request from the corresponding author.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-61-433-2018-supplement.
XS analyzed the data and wrote the paper; XW and YF performed the experiments; YM and DJ reviewed the paper; BH contributed to the sample collection; ZY conceived and designed the experiments.
The authors declare that they have no conflict of interest.
The research was financially supported by the National Plan of
863 Key Projects (2013AA102505-5), the Province Natural Scientific Foundation of
Yunnan, the project funded by the Priority Academic Program Development of
Jiangsu Higher Education Institutions (PAPD), and the Natural Science Foundation
of the Jiangsu Higher Education Institutions of China (17KJB230005).
Edited by: Steffen Maak
Reviewed by: Xianyong Lan, Runjun Yang,
and one anonymous referee
Allais, S., Journaux, L., Levéziel, H., Payet-Duprat, N., Raynaud, P., Hocquette, J.-F., Lepetit, J., Rousset, S., Denoyelle, C., and Bernard-Capel, C.: Effects of polymorphisms in the calpastatin and µ-calpain genes on meat tenderness in 3 French beef breeds, J. Anim. Sci., 89, 1–11, 2011.
Barendse, W., Harrison, B. E., Hawken, R. J., Ferguson, D. M., Thompson, J. M., Thomas, M. B., and Bunch, R. J.: Epistasis between calpain 1 and its inhibitor calpastatin within breeds of cattle, Genetics, 176, 2601–2610, 2007.
Buzanskas, M. E., Ventura, R. V., Chud, T. C. S., Bernardes, P. A., de Abreu Santos, D. J., de Almeida Regitano, L. C., de Alencar, M. M., de Alvarenga Mudadu, M., Zanella, R., and da Silva, M. V. G. B.: Study on the introgression of beef breeds in Canchim cattle using single nucleotide polymorphism markers, PloS One, 12, e0171660, https://doi.org/10.1371/journal.pone.0171660, 2017.
Calvo, J., Iguácel, L., Kirinus, J., Serrano, M., Ripoll, G., Casasús, I., Joy, M., Pérez-Velasco, L., Sarto, P., and Albertí, P.: A new single nucleotide polymorphism in the calpastatin (CAST) gene associated with beef tenderness, Meat Sci., 96, 775–782, 2014.
Casas, E., White, S., Wheeler, T., Shackelford, S., Koohmaraie, M., Riley, D., Chase, C., Johnson, D., and Smith, T.: Effects of and μ-markers in beef cattle on tenderness traits, J. Anim. Sci., 84, 520–525, 2006.
Castro, S., Ríos, M., Ortiz, Y., Manrique, C., Jiménez, A., and Ariza, F.: Association of single nucleotide polymorphisms in CAPN1, CAST and MB genes with meat color of Brahman and crossbreed cattle, Meat Sci., 117, 44–49, 2016.
Coll, F., McNerney, R., Guerra-Assunção, J. A., Glynn, J. R., Perdigão, J., Viveiros, M., Portugal, I., Pain, A., Martin, N., and Clark, T. G.: A robust SNP barcode for typing Mycobacterium tuberculosis complex strains, Nat. Commun., 5, 4812, https://doi.org/10.1038/ncomms5812, 2014.
Corva, P., Soria, L., Schor, A., Villarreal, E., Cenci, M. P., Motter, M., Mezzadra, C., Melucci, L., Miquel, C., and Paván, E.: Association of CAPN1 and CAST gene polymorphisms with meat tenderness in Bos taurus beef cattle from Argentina, Genet. Mol. Biol., 30, 1064–1069, 2007.
Curi, R. A., Chardulo, L. A. L., Mason, M., Arrigoni, M., Silveira, A. C., and De Oliveira, H.: Effect of single nucleotide polymorphisms of CAPN1 and CAST genes on meat traits in Nellore beef cattle (Bos indicus) and in their crosses with Bos taurus, Anim. Genet., 40, 456–462, 2009.
Davey, J. W., Hohenlohe, P. A., Etter, P. D., Boone, J. Q., Catchen, J. M., and Blaxter, M. L.: Genome-wide genetic marker discovery and genotyping using next-generation sequencing, Nat. Rev. Genet., 12, 499–510, 2011.
Enriquez-Valencia, C. E., Pereira, G. L., Malheiros, J. M., de Vasconcelos Silva, J. A. I., Albuquerque, L. G., de Oliveira, H. N., Chardulo, L. A. L., and Curi, R. A.: Effect of the g. 98535683A > G SNP in the CAST gene on meat traits of Nellore beef cattle (Bos indicus) and their crosses with Bos taurus, Meat Sci., 123, 64–66, 2017.
Gandolfi, G., Pomponio, L., Ertbjerg, P., Karlsson, A. H., Costa, L. N., Lametsch, R., Russo, V., and Davoli, R.: Investigation on CAST, CAPN1 and CAPN3 porcine gene polymorphisms and expression in relation to post-mortem calpain activity in muscle and meat quality, Meat Sci., 88, 694–700, 2011.
Johnston, D. J. and Graser, H. U.: Estimated gene frequencies of GeneSTAR markers and their size of effects on meat tenderness, marbling, and feed efficiency in temperate and tropical beef cattle breeds across a range of production systems, J. Anim. Sci., 88, 1917, https://doi.org/10.2527/jas.2009-2305, 2010.
Joo, S., Kim, G., Hwang, Y., and Ryu, Y.: Control of fresh meat quality through manipulation of muscle fiber characteristics, Meat Sci., 95, 828–836, 2013.
Kamruzzaman, M., ElMasry, G., Sun, D.-W., and Allen, P.: Non-destructive assessment of instrumental and sensory tenderness of lamb meat using NIR hyperspectral imaging, Food Chem., 141, 389–396, 2013.
Koohmaraie, M.: Biochemical factors regulating the toughening and tenderization processes of meat, Meat Sci., 43, 193–201, 1996.
Koohmaraie, M. and Geesink, G.: Contribution of postmortem muscle biochemistry to the delivery of consistent meat quality with particular focus on the calpain system, Meat Sci., 74, 34–43, 2006.
Lee, S.-H., Kim, S.-C., Chai, H.-H., Cho, S.-H., Kim, H.-C., Lim, D., Choi, B.-H., Dang, C.-G., Sharma, A., and Gondro, C.: Mutations in calpastatin and μ-calpain are associated with meat tenderness, flavor and juiciness in Hanwoo (Korean cattle): Molecular modeling of the effects of substitutions in the calpastatin/μ-calpain complex, Meat Sci., 96, 1501–1508, 2014.
Li, M., Sun, X., Hua, L., Lai, X., Lan, X., Lei, C., Zhang, C., Qi, X., and Chen, H.: SIRT1 gene polymorphisms are associated with growth traits in Nanyang cattle, Mol. Cell. Probes, 27, 215–220, 2013a.
Li, X., Ekerljung, M., Lundström, K., and Lundén, A.: Association of polymorphisms at DGAT1, leptin, SCD1, CAPN1 and CAST genes with color, marbling and water holding capacity in meat from beef cattle populations in Sweden, Meat Sci., 94, 153–158, 2013b.
Melo, S. V., Agnes, G., Vitolo, M. R., Mattevi, V. S., Campagnolo, P. D., and Almeida, S.: Evaluation of the association between the TAS1R2 and TAS1R3 variants and food intake and nutritional status in children, Genet. Mol. Biol., 40, 415–420, 2017.
Moeller, S., Miller, R., Edwards, K., Zerby, H., Logan, K., Aldredge, T., Stahl, C., Boggess, M., and Box-Steffensmeier, J.: Consumer perceptions of pork eating quality as affected by pork quality attributes and end-point cooked temperature, Meat Sci., 84, 14–22, 2010.
Nei, M. and Roychoudhury, A. K.: Sampling variances of heterozygosity and genetic distance, Genetics, 76, 379–390, 1974.
Renaville, B., Bacciu, N., Lanzoni, M., Corazzin, M., and Piasentier, E.: Polymorphism of fat metabolism genes as candidate markers for meat quality and production traits in heavy pigs, Meat Sci., 110, 220–223, 2015.
Sambrook, J. and Russell, D. W.: Molecular cloning: a laboratory manual. third, Cold pring Harbor Laboratory Press, New York, 2001.
Schenkel, F., Miller, S., Jiang, Z., Mandell, I., Ye, X., Li, H., and Wilton, J.: Association of a single nucleotide polymorphism in the calpastatin gene with carcass and meat quality traits of beef cattle, J. Anim. Sci., 84, 291–299, 2006.
Scollan, N. D., Dannenberger, D., Nuernberg, K., Richardson, I., MacKintosh, S., Hocquette, J.-F., and Moloney, A. P.: Enhancing the nutritional and health value of beef lipids and their relationship with meat quality, Meat Sci., 97, 384–394, 2014.
Sun, X.-M., Li, M.-X., Li, A.-M., Lan, X.-Y., Lei, C.-Z., Ma, W., Hua, L.-s., Wang, J., Hu, S.-R., and Chen, H.: Two novel intronic polymorphisms of bovine FGF21 gene are associated with body weight at 18 months in Chinese cattle, Livestock Sci., 155, 23–29, 2013.
Tait, R., Shackelford, S., Wheeler, T., King, D., Keele, J., Casas, E., Smith, T., and Bennett, G.: μ-Calpain, calpastatin, and genetic effects on preweaning performance, carcass quality traits, and residual variance of tenderness in a beef cattle population selected for haplotype and allele equalization, J. Anim. Sci., 92, 456–466, 2014.
Zhou, H., Byun, S., Frampton, C., Bickerstaffe, R., and Hickford, J.: Lack of association between CAST SNPs and meat tenderness in sheep, Anim. Genet., 39, 331–332, 2008.