the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Genomic characteristics of performance traits and adaptation to heat stress in desert Barki sheep
Adel M. Aboul-Naga
Hamdy Abdel-Shafy
Sherif Melak
Layaly Gamal
Shimaa H. Mustafa
Ibrahim Abousoliman
The Barki sheep is well adapted to desert conditions with intense solar radiation, low precipitation, and feed shortages. This review highlights advances in the genomic characterization of Barki sheep and outlines prospects for genetic improvement. Studies on the genomic architecture of their growth traits have identified multiple candidate genes, many of which overlap with those reported in other sheep breeds, suggesting the existence of shared biological pathways for growth regulation. Reproduction performance has been linked to genomic mutations in key loci, with limited evidence of polymorphisms in other genes, reflecting possible breed-specific selection pressures. Genomic studies have further revealed candidate genes related to milk production, although milk yield remains modest. Heat stress represents a major challenge for desert livestock, and the physiological responses of Barki sheep are evident through changes in the respiration rate, gas exchange, and breathing intensity. Candidate genes associated with heat tolerance are involved in multiple signaling and transduction pathways that regulate cellular and biochemical responses to heat stress, highlighting the strong adaptive capacity of the breed. Population-based genomic studies have shown that Barki sheep cluster closely with other Egyptian breeds but remain genetically distinct from exotic populations, underscoring their local adaptation. The considerable within-breed genetic variation provides opportunities for selection to improve lamb growth rates while maintaining moderate ewe size, lean meat, and heat resilience. Overall, genomic evidence indicates that Barki sheep represent a valuable model for climate-resilient livestock production. Owing to their unique adaptation to desert conditions and the availability of identified candidate genes for growth, reproductive performance, milk production, and heat tolerance, these genes provide a foundation for accurate genomic prediction. Future breeding programs should focus on their strengths to improve productivity while safeguarding their adaptive performance.
- Article
(432 KB) - Full-text XML
- BibTeX
- EndNote
Egyptian Barki desert sheep are widely distributed along the northwestern coast and represent an important source of income for Bedouin communities. The breed is primarily valued for its adaptability to harsh desert conditions, but it also has several economically important traits related to reproduction and meat production and quality (Aboulnaga and Abdelsabour, 2023). Barki sheep are named after the city of Barka in Libya and are spread across the coastal zone of the Western Desert (CZWD). They are raised under an extensive system; graze on natural rangelands and bushes sustained by limited rainfall; and follow a transhumant management pattern, moving to neighboring cultivated areas seeking feed and water. The breed is well adapted to arid and semiarid conditions, enduring high temperatures, intense solar radiation, low precipitation, and feed shortages. Compared to other Egyptian breeds, this is a fat-tailed, coarse-wool sheep with good fertility and mothering ability but relatively low growth performance (Fahmy et al., 1969). Barki sheep are thus characterized by their ability to survive, reproduce, and maintain productivity under harsh desert conditions (Aboul-Naga et al., 2022). There is a pressing need to understand the genetic and genomic basis of these adaptive traits to improve their performance sustainably.
The development of molecular genetics and its associated techniques have facilitated deeper insights into the genetic mechanisms underlying livestock traits. Different approaches, including candidate gene approaches, genome-wide association studies (GWASs), and selection signature analyses, have been used to identify quantitative trait loci (QTL) related to economically important traits. The candidate gene approach targets genes thought to be responsible for a given phenotype, whereas GWAS and selection signatures identify putative genes or chromosomal regions across the entire genome that influence trait variation (Beuzen et al., 2000).
Barki sheep have been the focus of several scientific investigations aiming to estimate genetic parameters for economically important traits (Melak et al., 2019; Sallam et al., 2019), to characterize genes or single-nucleotide polymorphisms (SNPs) associated with performance traits (Aboul-Naga et al., 2021; Abousoliman et al., 2020), and to assess their genetic diversity relative to other sheep breeds (Kim et al., 2016; Mwacharo et al., 2017). Despite these advances, knowledge gaps remain in elucidating the genomic architecture and biological pathways underlying their adaptation. Previous studies have identified several candidate genes associated with heat tolerance and productive traits. However, the border genomic mechanisms and regulatory networks supporting these adaptations remain insufficiently characterized. Specifically, limited attention has been given to the interactions among genes, the functional roles of identified variants in shaping their physiological responses, and the genomic basis of reproductive efficiency under chronic heat stress. Furthermore, most published work has relied on single-gene approaches, underscoring the need for system-level genomic studies that can capture the complexity of their adaptive responses.
The study of Barki sheep genomics holds both economic and ecological importance for production in arid and semiarid regions. Climate change projections indicate rising temperatures and expanding desertification, emphasizing the urgency of characterizing the genetic basis of heat tolerance. The Barki sheep represents a valuable genetic resource due to its ability to sustain productivity under high thermal loads, efficiently utilize limited feed resources, and support the livelihoods of the pastoral and smallholder communities in marginal environments. Clarifying these genomic mechanisms in the breed can directly help in identifying breeding and conservation strategies across comparable dryland systems.
From an economic perspective, understanding the genomic basis of these adaptations could facilitate the development of genomic selection programs aimed at enhancing productivity without compromising adaptive traits, support the design of breeding strategies to introduce heat tolerance into other populations, and strengthen conservation efforts targeting genetic diversity threatened by climate change. Ecologically, Barki sheep raised under a low-input production system offer a sustainable model for livestock rearing in fragile ecosystems, highlighting the potential to balance food security goals with environmental stewardship.
This review addresses the urgent need to link genomic knowledge with climate adaptation in livestock. Advancing our understanding of the genomic foundations of adaptive traits in Barki sheep makes it possible to design breeding programs that not only increase food security but also safeguard the genetic resources crucial for long-term resilience in challenging environments. The outcomes are directly relevant for communities in dryland regions worldwide that depend on livestock as a primary source of livelihood and nutrition.
Several studies have investigated the genetic basis of growth performance in Barki sheep via diverse approaches, such as whole-genome sequencing (WGS), partial sequencing, single-strand conformational polymorphism (SSCP), and kompetitive allele-specific polymerase chain reaction (KASP) genotyping (Table 1). Collectively, these studies have identified multiple genes associated with birth, weaning, and 6-month weights, including ADRβ3, CLPN3, PRKAG3, CLPN, LEP, MSTN, EYA2, GDF2, GDF10MEF2B, SLCI6A7, TBX15, IG3, TFAP2B, TNNC2, CPXM2, IGFBP3, and GH (Saleh et al., 2022). Among these genes, the ADRβ3 gene has received particular attention. In this context, Ibrahim (2014) demonstrated its association with growth traits such as marketing weight and body measurements in Barki sheep, which is consistent with earlier findings in New Zealand sheep breeds (Horrell et al., 2009; Forrest et al., 2003). Similarly, the CLPN3 gene was reported to influence the birth, weaning, and marketing weights of Barki lambs (Shehata et al., 2014). This finding aligns with broader evidence linking an SNP in intron 11 of the CLPN3 gene to birth weight across different sheep breeds (Chung et al., 2007) and to retail meat cuts (Bickerstaffe et al., 2008).
Other genes have also been highlighted for their roles in growth regulation. The PRKAG3 gene is associated with pre-weaning gain, marketing weight, the muscle index, and the mass index (Ibrahim, 2015), whereas the CLPN gene influences birth weight, final weight, and average daily gain (Mahrous et al., 2016). The leptin (LEP) gene has emerged as a strong candidate for growth-related traits, with polymorphisms significantly associated with weaning weight and pre-weaning average daily gain (Abousoliman et al., 2020). Its well-established role in regulating feed intake and energy metabolism further supports its biological relevance (Tahmoorespur et al., 2010; Choudhary et al., 2005).
More recently, WGS-based studies have expanded the list of candidate genes. Abousoliman et al. (2021a) detected several potential candidate genes associated with weaning weight and average daily gain in Barki lambs, such as EYA2, GDF2, GDF10, MEF2B, SLCI6A7, TBX15, TFAP2B, TNNC2, CPXM2, IG3, TFAP2B, TNNC2, and CPXM2. These genes are functionally linked to key biological processes, such as metabolism, growth, organ morphogenesis, skeletal muscle development, and cell proliferation and differentiation, highlighting their potential as molecular markers across different sheep breeds. Additional evidence was provided by Saleh et al. (2022) and Ibrahim et al. (2023), who reported associations between the IGFBP3, GH, HSP90, AB1HSF1, ST1P1, and ATP1A1 genes and growth traits including birth, weaning, 6-month weights, and average daily gain.
Taken together, these findings demonstrate that Barki lamb growth performance is influenced by a complex set of genes with pleiotropic roles in growth, metabolism, and muscle development. The convergence of evidence across studies and breeds underscores the potential of these loci as genetic markers for improving prediction strategies in Barki sheep.
Table 1Candidate genes studied for growth traits in Barki lambs.
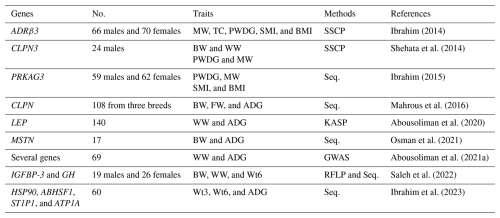
No.: number of animals; MW: marketing weight; TC: thigh circumference; PWDG: post-weaning daily gain; SMI: skeletal muscle index; BMI: body mass index; BW: birth weight; WW: weaning weight; FW: final weight; ADG: average daily gain; Wt3: weight at 3 months; Wt6: weight at 6 months; SSCP: single-strand conformation polymorphism; Seq.: sequencing; KASP: kompetitive allele-specific polymerase chain reaction; GWAS: genome-wide association study; RFLP: restriction fragment length polymorphism.
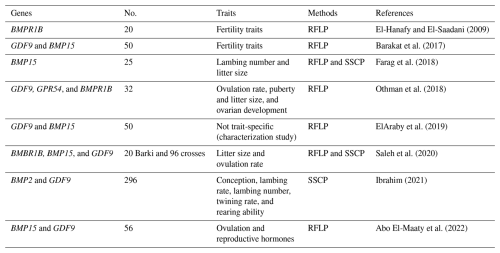
3.1 Reproductive performance
Barki ewes are well adapted to arid environments and are generally characterized by good fertility but low prolificacy (Gabr et al., 2016). Several studies have investigated genome variation related to reproductive traits (Table 2). DNA fragments of exon I and exon II of the GDF9 gene were analyzed, and no genetic polymorphisms were detected in some cases (ElAraby et al., 2019), whereas others reported genetic variation (Barakat et al., 2017) or Fec-GH mutation (Abo El-Maaty et al., 2022). Othman et al. (2018) confirmed the presence of the SNP c209G>A in the AG genotypes. Saleh et al. (2020) reported no Fec-GH mutation, although one amino acid substitution was detected (phenylalanine instead of isoleucine). Similarly, Ibrahim (2021) identified three SSCP banding patterns (C1, C2, and C3) with two SNPs (c25C>T and c260G>A), resulting in the amino acid substitutions p.Leu9Phe and p.Arg87His, respectively. These variants were significantly associated with the twining rate in Barki ewes (P<0.001), the number of lambs born (P<0.001), and the weight of lambs born (P<0.05). Fragments of exons I and II of the BMP15 gene revealed no genetic polymorphisms (ElAraby et al., 2019; Barakat et al., 2017) and no mutations in the FecXG locus (Abo El-Maaty et al., 2022; Saleh et al., 2020). However, Farag et al. (2018), via PCR-RFLP (restriction fragment length polymorphism), detected no polymorphisms at four loci (FecXB, FecXG, FecXH, and FecXI), whereas PCR-SSCP analysis revealed polymorphisms at three sites (FecXB, FecXG, and FecXH). Specifically, the AG at FecXB and the AC at FecXH were associated with improved twinning and increased lambing frequency in Barki ewes. No mutation was detected at the FecB locus of the BMBRIB gene (Othman et al., 2018; Saleh et al., 2020; El-Hanafy and El-Saadani, 2009). Analyses of exon II of the BMP2 gene revealed two variants (A1 and A2) with an SNP (c962A>T) leading to the substitution with p.His321Leu, which was significantly (P<0.05) associated with the number and weight of lambs weaned (Ibrahim, 2021). Additionally, exon V of the GPR54 gene included three genotypes (CC, CT, and TT) and one SNP (c100C>T) (Othman et al., 2018).
3.2 Milk production and composition
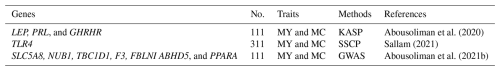
The milk yield of Barki sheep is generally lower than that of other Egyptian sheep breeds as no selection has been carried out for this trait in this desert breed. Notably, milk production and composition vary substantially among individuals, reflecting both genetic and environmental influences (Abousoliman et al., 2020). Using a candidate gene approach, Abousoliman et al. (2020) reported that SNP rs420693815 in exon 3 of LEP tended to increase milk yield and fat percentage, whereas rs422713690 in exon 3 of PRL had a close-to-significant effect (P=0.05) on milk yield. In addition, rs414991449 in exon 13 of GHRHR had a significant effect (P<0.001) on the total solids percentage (Table 3). Similar associations have been observed in other sheep breeds. For example, LEP genetic polymorphisms are significantly associated with milk yield in Najdi desert sheep in Saudi Arabia (Mahmoud et al., 2014). Similarly, genetic polymorphisms in PRL have been linked to milk production in Serra da Estrella and East Friesian sheep (Ramos et al., 2009; Moioli et al., 2007). The GHRHR gene mediates the effects of its ligand, growth-hormone-releasing hormone (GHRH), thereby regulating growth hormone (GH) synthesis and secretion (Giustina and Veldhuis, 1998).
In addition to these classical milk-related genes, immune system genes have also emerged as relevant candidates. Sallam (2021) detected a mutation (rs592076818; c1710C>A) in the coding sequence of the toll-like receptor 4 (TLR4) gene, resulting in an amino acid substitution (p. Asn570Lys) that had a significant effect (P<0.05) on milk fat and protein percentages, as well as a close-to-significant effect on daily milk yield (Table 3). Consistently with these findings, several studies have reported significant associations between genetic polymorphisms of TLR4 and milk production traits in cattle (Sharma et al., 2006; Beecher et al., 2010; Noori et al., 2013; Zhou et al., 2017). Using genome scanning, Abousoliman et al. (2021b) reported potential candidate genes associated with milk yield and milk composition, including SLC5A8, NUB1, TBC1D1, KLF3, ABHD5, PPARA, and FBLN1 (Table 3). These genes have also been identified as markers for milk production, milk composition, and mammary gland development in different livestock species, highlighting their broader biological importance.
Barki sheep exposed to heat stress (HS) presented a significant increase in respiration rate (RR) and minute ventilation volume (MVV) and a decrease in tidal volume (TV) and gas exchange (shallow, rapid panting) (Elbeltagy et al., 2015b). Changes in RR and gas volume were more pronounced in terms of the incidence of deep breaths observed, whereas changes in thermal parameters were less detectable. Within the breed, medium-sized desert sheep appeared to be more compatible with the prevailing hot and dry conditions, highlighting possible interbreed variations in terms of heat adaptation. Consistent results were reported for Barki populations raised at different locations in the CZWD (Table 4).
Table 4Effects of climate change on the physiological parameters of Barki sheep.
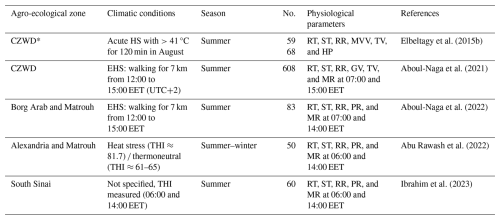
* CZWD: coastal zone of Western Desert; No: number of animals; HS: heat stress; EHS: exercise heat stress; ST: skin temperature; RR: respiration rate; MVV: minute ventilation volume; TV: tidal volume; GV: gas volume; MR: metabolic rate; PR: production rate; HP: heat production.
Table 5Candidate genes and genomic studies for tolerance to heat stress in Barki sheep.
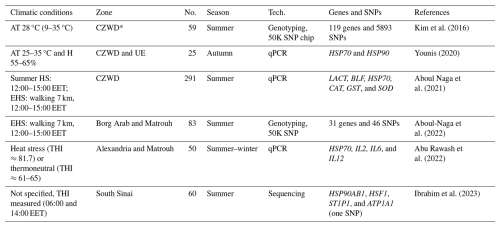
* CZWD: coastal zone of the Western Desert; no.: number of animals; UE: Upper Egypt; AT: air temperature; H: humidity; HS: heat stress; EHS: exercise heat stress; SNP: single-nucleotide polymorphism; qPCR: quantitative PCR.
At the genomic level, increasing evidence from several studies has highlighted the genetic variants and candidate genes that contribute to the remarkable adaptation of Barki sheep to HS (Table 5). Kim et al. (2016) identified 18 genes in two regions on chromosomes 6 and 12, each spanning one gene, CSN3 and PCDH9, respectively. In addition, seven candidate regions were identified across chromosomes 1, 3, 7, 10, 12, 13, 17, 19, and 21. Two of these regions, located on chromosomes 3 and 13, each contained a single gene (TRHDE and BMP2, respectively). In total, 119 genes were detected, many of which are involved in multiple signaling and signal transduction pathways regulating diverse cellular and biochemical processes. Genes under selection were linked to traits such as thermotolerance (FGF2, GNAI3, and PLCB1), body size and development (BMP2, BMP4, GJA3, and GJB2), energy and digestive metabolism (MYH, TRHDE, and ALDH1A3), and nervous and autoimmune responses (GRIA1, IL2, IL7, IL21, and IL1R1). Moreover, eight candidate genes with selection signatures were identified on a conserved syntenic segment on chromosome 10, providing strong evidence for natural selection acting in the Barki environment. Gene expression studies further support these genomic findings. In this context, Younis (2020) revealed that the expression levels of the HSP70 and HSP90 genes were downregulated in Barki sheep but upregulated in Aboud Leik southern local sheep. They recommended the use of HSP70 and HSP90 expression profiles as reference markers for selecting animals with improved adaptability in arid conditions.
Similarly, Aboul Naga et al. (2021) studied six genes associated with oxidative stress responses in different farm animals. The expression levels of these genes differed between low-tolerance (LT) and high-tolerance (HT) Barki sheep under exercise heat stress (EHS). While the SOD gene was expressed at lower levels in HT sheep, other genes (GST, CAT, HSP70, BLF, and LACT) were expressed at higher levels in HT animals than in LT animals. Aboul-Naga et al. (2022) reported that 46 SNPs are significantly associated (P≤0.001) with heat tolerance. These SNPs, located mainly between OAR7_60745094.1 and OAR7_60704536.1, had effects ranging from 0.57 to 1.47 units, either positive or negative. Gene ontology analysis revealed that the associated genes participate in calcium and manganese binding and/or transport, epidermal growth, and cell adhesion, with additional roles for epidermal growth factor (EGF)-domain-related genes, kinases, growth, and collagen-containing extracellular matrix proteins. Among the most significant SNPs were variants in the MYO5A, PRKG1, GSTCD, and RTN1 genes (P≤0.0001). Notably, MYO5A encodes a protein widely distributed in the melanin-producing neural crest of the skin. Further associations identified SNPs in the PLCB1, STEAP3, KSR2, UNC13C, PEBP4, and GPAT2 genes. The involvement of genes linked to pigmentation and tail fat deposition suggests a functional basis for why near-eastern sheep breeds outperform temperate breeds under HS in hot, dry environments. These findings further imply that climate change may drive shifts in livestock distribution in arid regions, favoring sheep over cattle and goats over sheep, highlighting the importance of utilizing local breeds naturally selected under desert conditions.
Additional evidence emphasizes the role of season-specific responses. Abu Rawash et al. (2022) reported nonsignificant seasonal differences in tolerance gene expression in Barki sheep. However, they reported summer upregulation of IL2 and IL6 alongside winter upregulation of HSP70, suggesting distinct roles; HSP70 is a molecular marker for cold adaptation, and IL2 and IL6 are involved in heat adaptation. These results illustrate that Barki sheep withstand HS by adjusting their physiological parameters while maintaining stable blood plasma constituents. In addition, Ibrahim et al. (2023) investigated the associations between Barki lamb performance and nucleotide sequence variations in growth- and efficiency-related genes (GB1CAST, GB2LEP, GB1MYLK4, GB1MEF2B, and GB2TRPV1). The SNPs identified for growth were strongly correlated with heat tolerance, indicating that selection for growth and heat tolerance may be achieved simultaneously. They recommended exploiting this genetic variance as a proxy marker for breeding Barki sheep resilient to both environmental stress and production demands.
Table 6Genomic diversity studies in Barki sheep.
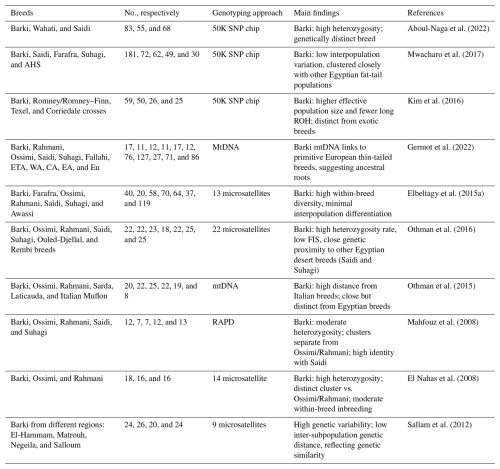
No.: number of animals; ETA: eastern tropical African breeds; WA: western Asian fat-tail breeds; CA: central Asian breeds; EA: eastern Asian breeds; Eu: European breeds; mtDNA: mitochondrial DNA; RAPD: random amplified polymorphic DNA; ROH: runs of homozygosity.
Genomic diversity in Egyptian sheep has been investigated via both genomic and microsatellite markers, providing complementary perspectives on population structure. Studies based on genome-wide SNP data (Kim et al., 2016; Mwacharo et al., 2017; Aboul-Naga et al., 2022) revealed that Egyptian breeds tend to cluster closely together and remain clearly separated from the exotic breeds (Table 6). Within this broader pattern, Egyptian sheep were more closely related to western Asian fat-tailed breeds than to European breeds. The Barki breed was distinguished from other Egyptian populations, reflecting its unique genetic background.
Microsatellite-based studies (Elbeltagy et al., 2015a; Mahfouz et al., 2008; El Nahas et al., 2008; Sallam et al., 2012; Othman et al., 2015; Othman et al., 2016; Germot et al., 2022) largely support these findings while offering a finer resolution in terms of the relationships among local breeds. These studies consistently reported two main clusters: one comprising Ossimi and Rahmani sheep and another divided into two subclusters, with Barki forming one branch and Saidi and Suhagi forming the other. This pattern indicated that Barki sheep are genetically closer to Saidi and Suhagi than to Ossimi and Rahmani, which are closely related to each other. Within the Barki population, subpopulation analysis revealed two genetic groups: Hammam and Negila on one side and Matrouh and Salloum on the other. Despite these subdivisions, the overall genetic distances between subpopulations were limited, reflecting a good degree of genetic similarity. Together, these findings illustrate how both genome-wide and microsatellite approaches converge into a consistent picture of Egyptian sheep diversity while also highlighting the unique genetic profile of the Barki breed.
This review highlights the unique adaptive capacity of Barki sheep and combines current knowledge on their productive performance, physiological resilience, and genomic potential. Previous studies have largely provided descriptive accounts of breed characteristics; the combination presented here emphasizes the strategic role of Barki sheep in supporting livestock production in arid and semiarid regions. Despite their importance, research on Barki sheep remains fragmented, with limited genomic resources and few efficient evaluations of productivity–resilience trade-offs. Addressing these gaps is critical for designing effective breeding and conservation strategies. Integrating genomic tools with traditional selection schemes offers a promising pathway to accelerate genetic improvement while preserving adaptive traits that safeguard survival under climate stress. The evidence summarized in this review underscores the need for coordinated efforts to establish and expand the reference population, refine genomic prediction approaches tailored to performance and adaptive traits, and implement community-based breeding programs. These initiatives would directly strengthen the sustainable improvement of Barki sheep and offer transferable strategies for enhancing the resilience of other indigenous breeds in arid production systems.
No data sets were used in this article.
AMAN contributed to the conception and design of the work and wrote the first draft of the paper. SHM, SHH, LG, and ISH organized the database and reviewed the literature. HA contributed to the writing and revision of the paper.
The contact author has declared that none of the authors has any competing interests.
No ethical consent was required for this review.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors. Views expressed in the text are those of the authors and do not necessarily reflect the views of the publisher.
The authors would like to thank members of the “Molecular Genetic Scientific Group – MGSG” of APRI for their valuable advice and feedback on the paper.
This paper was edited by Steffen Maak and reviewed by Sameh A. Abdelnour and one anonymous referee.
Abo El-Maaty, A. M., Abdelhafez, M. A., and Mahrous, K. F.: Effect of diet restriction and polymorphism of bone morphogenetic protein-15 and growth differentiation factor-9 (GDF9) on reproductive performance of three Egyptian fat tail sheep breeds, J. Adv. Vet. Res., 12, 18–24, 2022.
Aboul Naga, A. M., Abdel Khalek, T. M., Osman, M., Elbeltagy, A. R., Abdel-Aal, E. S., Abou-Ammo, F. F., and El-Shafie, M. H.: Physiological and genetic adaptation of desert sheep and goats to heat stress in the arid areas of Egypt, Small Rumin. Res., 203, 106499, https://doi.org/10.1016/j.smallrumres.2021.106499, 2021.
Aboul-Naga, A., Elshafie, M., Khalifa, H., Osman, M., Abdel Khalek, T., El-Beltagi, A., Abdel-Aal, E., Abdel-Sabour, T., Rekik, M., Rouatbi, M., and Rischowesky, B.: Tolerance capability of desert sheep and goats to exercise heat stress under hot dry conditions, and its correlation with their production performance, Small Rumin. Res., 205, 106550, https://doi.org/10.1016/j.smallrumres.2021.106550, 2021.
Aboul-Naga, A. M., Alsamman, A. M., El Allali, A., Elshafie, M. H., Abdelal, E. S., Abdelkhalek, T. M., Abdelsabour, T. H., Mohamed, L. G., and Hamwieh, A.: Genome-wide analysis identified candidate variants and genes associated with heat stress adaptation in Egyptian sheep breeds, Front. Genet., 13, 898522, https://doi.org/10.3389/fgene.2022.898522, 2022.
Aboulnaga, A. and Abdelsabour, T.: Review of sheep and goat Research and Development in Egypt since the forties: II-phenotypic characteristics, production, and reproduction performance of local sheep breeds, J. Anim. Poult. Prod., Mansoura Univ., 14, 43–52, https://doi.org/10.21608/jappmu.2023.206649.1073, 2023.
Abousoliman, I., Reyer, H., Oster, M., Muráni, E., Mourad, M., Abdel-Salam Rashed, M., Mohamed, I., and Wimmers, K.: Analysis of candidate genes for growth and milk performance traits in the Egyptian Barki sheep, Animals (Basel), 10, 197, https://doi.org/10.3390/ani10020197, 2020.
Abousoliman, I., Reyer, H., Oster, M., Murani, E., Mohamed, I., and Wimmers, K.: Genome-wide analysis for early growth-related traits of the locally adapted Egyptian Barki sheep, Genes (Basel), 12, 1243, https://doi.org/10.3390/genes12081243, 2021a.
Abousoliman, I., Reyer, H., Oster, M., Murani, E., Mohamed, I., and Wimmers, K.: Genome-Wide SNP analysis for milk performance traits in indigenous sheep: A case study in the Egyptian Barki sheep, Animals (Basel), 11, 1671, https://doi.org/10.3390/ani11061671, 2021b.
Abu Rawash, R. A., Sharaby, M. A., Hassan, G. E.-D. A., Elkomy, A. E., Hafez, E. E., Hafsa, S. H. A., and Salem, M. M.: Expression profiling of HSP 70 and interleukins 2, 6 and 12 genes of Barki sheep during summer and winter seasons in two different locations, Int. J. Biometeorol., 66, 2047–2053, https://doi.org/10.1007/s00484-022-02339-6, 2022.
Barakat, I. A., Salem, L. M., Daoud, N. M., Khalil, W. K., and Mahrous, K. F.: Genetic polymorphism of candidate genes for fecundity traits in Egyptian sheep breeds, Biomed. Res., 28, 851–857, 2017.
Beecher, C., Daly, M., Childs, S., Berry, D. P., Magee, D. A., McCarthy, T. V., and Giblin, L.: Polymorphisms in bovine immune genes and their associations with somatic cell count and milk production in dairy cattle, BMC Genet., 11, 99, https://doi.org/10.1186/1471-2156-11-99, 2010.
Beuzen, N., Stear, M., and Chang, K.: Molecular markers and their use in animal breeding, Vet. J., 160, 42–52, https://doi.org/10.1053/tvjl.2000.0468, 2000.
Bickerstaffe, R., Hickford, J., Gately, K., and Zhou, H.: Association of polymorphic variations in calpastatin with meat tenderness and yield of retail meat cuts in lambs, Proceedings of international congress of meat science, 1–3, Helsinki, Finland, 2008.
Choudhary, V., Kumar, P., Bhattacharya, T. K., Bhushan, B., and Sharma, A.: DNA polymorphism of leptin gene in Bos indicus and Bos taurus cattle, Genet. Mol. Biol., 28, 740–742, https://doi.org/10.1590/S1415-47572005000500014, 2005.
Chung, H., Choi, B., Jang, G., Lee, K., Kim, H., Yoon, S., Im, S., Davis, M., and Hines, H.: Effect of variants in the ovine skeletal-muscle-specific calpain gene on body weight, J. Appl. Genet., 48, 61–68, 2007.
ElAraby, I., Magdy, S., Awad, A., and Saleh, A.: Polymorphisms of growth differentiation factor 9 (GDF 9) and bone morphogenetic protein 15 (BMP15) genes in Barki and Rahmani sheep breeds, Zag. Vet. J., 47, 160–167, https://doi.org/10.21608/zvjz.2019.10071.1025, 2019.
Elbeltagy, A. R., Aboul-Naga, A., Hassen, H., Rischkowsky, B., and Mwacharo, J. M.: Genetic diversity and structure in Egyptian indigenous sheep populations mirror patterns of anthropological interactions, Small Rumin. Res., 132, 137–142, https://doi.org/10.1016/j.smallrumres.2015.10.020, 2015a.
Elbeltagy, A., Aboul-Naga, A., Khalifa, H., Abdel Khalek, T., Elshafie, M., and Rischovesky, B.: Biological and mathematical analysis of desert sheep and goats responses to natural and acute heat stress, in Egypt, Egypt. J. Anim. Prod., 52, 45–52, 2015b.
El-Hanafy, A. A. and El-Saadani, M.: Fingerprinting of FecB gene in five Egyptian sheep breeds, Biotechnol. Anim. Husb., 25, 205–212, https://doi.org/10.2298/BAH0904205E, 2009.
El Nahas, S. M., Hassan, A. A., Mossallam, A., Mahfouz, E. R., Bibars, M. A., Oraby, A., and De Hondt, H.: Analysis of genetic variation in different sheep breeds using microsatellites, Afr. J. Biotechnol., 7, 1060–1068, 2008.
Fahmy, M., Galal, E. S. E., Ghanem, Y., and Khishin, S.: Genetic parameters of Barki sheep raised under semi-arid conditions, Anim. Sci., 11, 361–367, https://doi.org/10.1017/S0003356100027008, 1969.
Farag, I., Aboelhassan, D., Ghaly, I., Darwish, H., Darwish, A., and Eshak, M.: Effect of genetic polymorphism of BMP15 gene on improving twin production in Egyptian small ruminants, Res. J. Anim. Vet. Sci., 10, 6–14, https://doi.org/10.22587/rjavs.2018.10.1.2, 2018.
Forrest, R. H., Hickford, J. G., Hogan, A., and Frampton, C.: Polymorphism at the ovine beta3-adrenergic receptor locus: associations with birth weight, growth rate, carcass composition and cold survival, Anim. Genet., 34, 19–25, https://doi.org/10.1046/j.1365-2052.2003.00936.x, 2003.
Gabr, A., Shalaby, N., and Ahmed, M.: Effect of ewe born type, growth rate and weight at conception on the ewe subsequent productivity of Rahmani sheep, Asian. J. Anim. Vet. Adv., 11, 732–736, https://doi.org/10.3923/ajava.2016.732.736, 2016.
Germot, A., Khodary, M. G., Othman, O. E.-M., and Petit, D.: Shedding Light on the Origin of Egyptian Sheep Breeds by Evolutionary Comparison of Mitochondrial D-Loop, Animals (Basel), 12, 2738, https://doi.org/10.3390/ani12202738, 2022.
Giustina, A. and Veldhuis, J. D.: Pathophysiology of the neuroregulation of growth hormone secretion in experimental animals and the human, Endocr. Rev., 19, 717–797, https://doi.org/10.1210/edrv.19.6.0353, 1998.
Horrell, A., Forrest, R., Zhou, H., Fang, Q., and Hickford, J.: Association of the ADRB3 gene with birth weight and growth rate to weaning in New Zealand Romney sheep, Anim. Genet., 40, 251–254, https://doi.org/10.1111/j.1365-2052.2008.01807.x, 2009.
Ibrahim, A.: Single strand conformational polymorphism of ADRß3 gene and its association with live performance traits in Barki sheep, Egypt. J. Genet. Cytol., 43, 287–299, 2014.
Ibrahim, A.: The PRKAG3 gene polymorphisms and their associations with growth performance and body indices in Barki lambs, Egypt. J. Sheep Goats Sci., 10, 1–13, 2015.
Ibrahim, A. H.: Genetic variants of the BMP2 and GDF9 genes and their associations with reproductive performance traits in Barki ewes, Small Rumin. Res., 195, 106302, https://doi.org/10.1016/j.smallrumres.2020.106302, 2021.
Ibrahim, S., Al-Sharif, M., Younis, F., Ateya, A., Abdo, M., and Fericean, L.: Analysis of potential genes and economic parameters associated with growth and heat tolerance in sheep (Ovis aries), Animals (Basel), 13, 353, https://doi.org/10.3390/ani13030353, 2023.
Kim, E.-S., Elbeltagy, A. R., Aboul-Naga, A., Rischkowsky, B., Sayre, B., Mwacharo, J. M., and Rothschild, M. F.: Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment, Heredity, 116, 255–264, https://doi.org/10.1038/hdy.2015.94, 2016.
Mahfouz, E. R., Othman, O. E., El Nahas, S. M., and El Barody, M.: Genetic variation between some Egyptian sheep breeds using RAPD-PCR, Res. J. Cell Mol. Biol., 2, 46–52, 2008.
Mahmoud, A., Saleh, A., Almealamah, N., Ayadi, M., Matar, A., Abou-Tarboush, F., Aljumaah, R., and Abouheif, M.: Polymorphism of leptin gene and its association with milk traits in Najdi sheep, J. Pure Appl. Microbio., 8, 2953–2959, 2014.
Mahrous, K., Hassanane, M., Shafey, H., Abdel Mordy, M., and Rushdi, H.: Association between single nucleotide polymorphism in ovine Calpain gene and growth performance in three Egyptian sheep breeds, J. Genet. Eng. Biotechnol., 14, 233–240, https://doi.org/10.1016/j.jgeb.2016.09.003, 2016.
Melak, S., Mansour, H., Aboul-Naga, A., Osman, M. A., Elbeltagy, A., and El Sayed, M.: Genetic parameters for growth performance traits of Egyptian Barki lambs using random regression model, Arab Univ. J. Agric. Sci., 27, 383–393, https://doi.org/10.21608/ajs.2019.43577, 2019.
Moioli, B., D'Andrea, M., and Pilla, F.: Candidate genes affecting sheep and goat milk quality, Small Rumin. Res., 68, 179–192, https://doi.org/10.1016/j.smallrumres.2006.09.008, 2007.
Mwacharo, J. M., Kim, E.-S., Elbeltagy, A. R., Aboul-Naga, A. M., Rischkowsky, B. A., and Rothschild, M. F.: Genomic footprints of dryland stress adaptation in Egyptian fat-tail sheep and their divergence from East African and western Asia cohorts, Sci. Rep., 7, 17647, https://doi.org/10.1038/s41598-017-17775-3, 2017.
Noori, R., Mahdavi, A., Edriss, M., Rahmani, H., Talebi, M., and Soltani-Ghombavani, M.: Association of polymorphism in Exon 3 of toll-like receptor 4 gene with somatic cell score and milk production traits in Holstein dairy cows of Iran, S. Afr. J. Anim. Sci., 43, 493–498, https://doi.org/10.4314/sajas.v43i4.6, 2013.
Osman, N. M., Shafey, H. I., Abdelhafez, M. A., Sallam, A. M., and Mahrous, K. F.: Genetic variations in the Myostatin gene affecting growth traits in sheep, Vet. World, 14, 475–482, https://doi.org/10.14202/vetworld.2021.475-482, 2021.
Othman, O., Abd El-Kader, H., Abd El-Rahim, A., Abd El-Moneim, O., and Alam, S.: Genetic characterization of three genes associated with fertility performance in Egyptian small ruminant breeds, Indian J. Anim. Sci., 88, 200–205, https://doi.org/10.56093/ijans.v88i2.79336, 2018.
Othman, O. E., Pariset, L., Balabel, E. A., and Marioti, M.: Genetic characterization of Egyptian and Italian sheep breeds using mitochondrial DNA, J. Genet. Eng. Biotechnol., 13, 79–86, https://doi.org/10.1016/j.jgeb.2014.12.005, 2015.
Othman, O. E. M., Payet-Duprat, N., Harkat, S., Laoun, A., Maftah, A., Lafri, M., and Da Silva, A.: Sheep diversity of five Egyptian breeds: Genetic proximity revealed between desert breeds: Local sheep breeds diversity in Egypt, Small Rumin. Res., 144, 346–352, https://doi.org/10.1016/j.smallrumres.2016.10.020, 2016.
Ramos, A., Matos, C., Russo-Almeida, P., Bettencourt, C., Matos, J., Martins, A., Pinheiro, C., and Rangel-Figueiredo, T.: Candidate genes for milk production traits in Portuguese dairy sheep, Small Rumin. Res., 82, 117–121, https://doi.org/10.1016/j.smallrumres.2009.02.007, 2009.
Saleh, A. A., Hammoud, M., Dabour, N. A., Hafez, E., and Sharaby, M. A.: BMPR-1B, BMP-15 and GDF-9 genes structure and their relationship with litter size in six sheep breeds reared in Egypt, BMC Res. Notes, 13, 215, https://doi.org/10.1186/s13104-020-05047-9, 2020.
Saleh, A. A., Hammoud, M. H., Dabour, N. A., Hafez, E. E. E., and Sharaby, M. A.: Genetic variability in GH and IGFBP-3 genes and their association with growth performance in Egyptian sheep, Appl. Vet. Res., 1, e2022012, https://doi.org/10.31893/avr.2022012, 2022.
Sallam, A., Galal, S., Rashed, M., and Alsheikh, S.: Genetic diversity in Barki sheep breed in its native tract in Egypt, Egypt. J. Anim. Prod., 49, 19–28, 2012.
Sallam, A. M.: A missense mutation in the coding region of the toll-like receptor 4 gene affects milk traits in Barki sheep, Anim. Biosci., 34, 489–498, https://doi.org/10.5713/ajas.19.0989, 2021.
Sallam, A. M., Ibrahim, A. H., and Alsheikh, S. M.: Genetic evaluation of growth in Barki sheep using random regression models, Trop. Anim. Health Prod., 51, 1893–1901, https://doi.org/10.1007/s11250-019-01885-3, 2019.
Sharma, B., Leyva, I., Schenkel, F., and Karrow, N.: Association of toll-like receptor 4 polymorphisms with somatic cell score and lactation persistency in Holstein bulls, J. Dairy Sci., 89, 3626–3635, https://doi.org/10.3168/jds.S0022-0302(06)72402-X, 2006.
Shehata, M., Ismail, I., and Ibrahim, A.: Variation in exon 10 of the ovine calpain3 gene and its association with growth and carcass traits in Egyptian Barki lambs, Egypt. J. Genet. Cytol., 43, 231–240, 2014.
Tahmoorespur, M., Taheri, A., Valeh, M. V., Saghi, D. A., and Ansary, M.: Assessment relationship between leptin and ghrelin genes polymorphisms and estimated breeding values (EBVs) of growth traits in Baluchi sheep, J. Anim. Vet. Adv., 9, 2460–2465, https://doi.org/10.3923/javaa.2010.2460.2465, 2010.
Younis, F.: Expression pattern of heat shock protein genes in sheep, Mansoura Vet. Med. J., 21, 1–5, https://doi.org/10.21608/mvmj.2020.21.001, 2020.
Zhou, H., Cheng, L., Gong, H., Byun, S. O., Edwards, G. R., and Hickford, J. G.: Variation in the Toll-like Receptor 4 (TLR4) gene affects milk traits in dairy cows, J. Dairy Res., 84, 426–429, https://doi.org/10.1017/S0022029917000711, 2017.
- Abstract
- Introduction
- Genomic analysis of Barki lamb performance traits
- Genomic analysis of Barki ewes' performance traits
- Adaptation of Barki sheep to heat stress under desert conditions
- Genomic diversity
- Conclusions
- Data availability
- Author contributions
- Competing interests
- Ethical statement
- Disclaimer
- Acknowledgements
- Review statement
- References
- Abstract
- Introduction
- Genomic analysis of Barki lamb performance traits
- Genomic analysis of Barki ewes' performance traits
- Adaptation of Barki sheep to heat stress under desert conditions
- Genomic diversity
- Conclusions
- Data availability
- Author contributions
- Competing interests
- Ethical statement
- Disclaimer
- Acknowledgements
- Review statement
- References