the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Polymorphisms of the BMPR1B, BMP15 and GDF9 fecundity genes in four Chinese sheep breeds
Jinxin Wang
Yufang Liu
Siwu Guo
Ran Di
Xiangyu Wang
Xiaoyun He
Mingxing Chu
Numerous studies on prolific sheep breeds have shown that the transforming growth factor beta (TGF-β) superfamily members, including bone morphogenetic protein receptor type 1B (BMPR1B), bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9), are the essential regulators of ovulation rate and litter size. In total, 11 known mutations (1 in BMPR1B, 6 in BMP15 and 4 in GDF9) are able to increase the ovulation rate and litter size, respectively. In this study, the genomic DNA was isolated from 512 high-prolificacy sheep (including the Small-tailed Han, Hu and Wadi sheep breeds) and 164 low-prolificacy sheep (Tan sheep), and genotyping of the specific mutations of the three fecundity-related genes was carried out by sequencing. The results showed that the FecB mutation in BMPR1B was detected in all four sheep breeds, and the frequency of B allele was significantly higher in the high-prolificacy breeds than that in the low-prolificacy breed (P<0.001). A novel mutation, c.T755C (named S1), was found in BMP15 from the four sheep breeds. However, known mutations such as FecXI, FecXH, FecXB, FecXG, FecXL and FecXR were not detected in these breeds. Three known loci (G1, G3 and G4) and a new mutation, c.A1515G (named S2), were found in GDF9, and the other three known mutations (FecGH (G8), FecGE and FecTT) were not detected in all four sheep breeds. The genotype distribution at the G1 and G4 loci had significant differences between the low-prolificacy sheep breed and the other three high-prolificacy sheep breeds. There was no difference in the genotype distribution at the G1 and G4 loci between the three high-prolificacy sheep breeds. Haplotype analysis of the four polymorphic loci in GDF9 suggested that H4 (GGAA) was the preponderant haplotype in the three high-prolificacy sheep breeds, but H1 (GGGG) was in the low-prolificacy sheep breed. These results preliminarily showed that the BMPR1B and GDF9 might be major genes influencing the prolificacy of Chinese sheep breeds.
- Article
(2415 KB) - Full-text XML
- BibTeX
- EndNote
Many studies have shown that the reproductive performance of sheep is affected by genetic variation. Finding polymorphism of genes can provide potential molecular genetic markers for sheep reproduction. This study investigated the polymorphisms of three major reproductive genes (BMPR1B, BMP15 and GDF9) in Chinese sheep and found that BMPR1B and GDF9 might be the major genes influencing the reproductive performance of Chinese sheep breeds. This result might contribute to improving the prolificacy traits of Chinese native sheep in the future.
Ovulation rate and litter size are important reproduction traits in sheep and are of high economic value. Reproduction traits typically have low to medium heritability and do not exhibit a noticeable response to phenotypic selection. Therefore, inclusion of genetic information of the genes associated with reproductive ability could efficiently enhance the selection response (Abdoli et al., 2016). In sheep, genetic variation in litter size and ovulation rate has been widely documented, and many findings have indicated that substantial differences between breeds and within breeds occurred in different mechanisms. Three members of the transforming growth factor-β (TGF-β) superfamily, i.e., bone morphogenetic protein receptor type 1B (BMPR1B), bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9), have been shown to be essential regulators for ovulation rate and follicular development (Davis, 2005). GDF9 and BMP15 are co-expressed exclusively in oocytes throughout most of the folliculogenesis and play central roles in controlling ovarian physiology (Stocker et al., 2020).
The BMPR1B known as FecB on sheep chromosome 6 includes 16 exons (gene ID 443454). The A–G transition was detected at nucleotide 746 of BMPR1B cDNA, causing a nonsynonymous substitution of glutamine with an arginine (Q249R) at position 249 of the mature protein in Booroola Merino sheep (Mulsant et al., 2001; Souza et al., 2001; Wilson et al., 2001). The FecBB allele leads to an additive effect for the ovulation rate and an increased litter size. The genotypes of FecB mutation in the ewes have been classified as homozygous carriers (BB) with more than five ovulations per estrous cycle, heterozygous carriers (B+) with three to four ovulations and homozygous non-carriers () with an ovulation rate of two or less (Davis et al., 1982). A recent study of Small-tailed Han sheep showed that BMPR1B was highly expressed in hypothalamus, ovary, uterus and oviduct tissue during the follicular phase, and BMPR1B was expressed significantly more in the hypothalamus of polytocous ewes than in monotocous ewes during both the follicular and luteal phases (Wen et al., 2021). The BMP15 known as FecX is on sheep chromosome X, and its coding sequence contains two exons (Galloway et al., 2000). Eight mutations have been detected within the sheep BMP15, including FecXI (Inverdale, V299D), FecXH (Hanna, Q291Ter), FecXB (Belclare, S367I), FecXG (Galway, Q239Ter), FecXL (Lacaune, C321Y), FecXR (Rasa Aragonesa, 154-159del WVQKSP), FecXO and FecXGr (Galloway et al., 2000; Hanrahan et al., 2004; Bodin et al., 2007; Martinez-Royo et al., 2008; Monteagudo et al., 2009; Abdoli et al., 2016; Demars et al., 2013). There are 13 identified variations in the Chinese Luzhong mutton sheep breed, of which 6 are novel (Di et al., 2021). All these mutations led to increased ovulation rates in the heterozygous animals and sterility in the homozygous animals. As a close homolog of BMP15, the mutations of GDF9 also result in similar genotypes to those in BMP15, and they cause an increased ovulation rate in heterozygous ewes, while homozygotes are sterile (Hanrahan et al., 2004; McNatty et al., 2005a). The GDF9 known as FecG is on sheep chromosome 5 and contains two exons. Eight single-nucleotide polymorphisms (SNPs), labeled G1–G8, have been identified in the entire coding region of the sheep GDF9 (Hanrahan et al., 2004). Four mutations in GDF9 have been identified as causing either infertility or an increased ovulation rate, including G8 (FecGH) (Belclare and Cambridge, S395F) (Hanrahan et al., 2004), FecGE (Santa Ines, F345C) (Melo et al., 2008; Silva et al., 2011) and FecTT (Thoka, S427R) (Nicol et al., 2009). Two novel SNPs of GDF9 (g.41768501A > G and g.41768485 G > A) were found in Luzhong mutton sheep and may be potential genetic markers for improving litter size (Wang et al., 2021).
Small-tailed Han sheep, Hu sheep and Wadi sheep are excellent native breeds in China for their significant characteristics of year-round estrus and hyper-prolificacy. Tan sheep is a native breed in China with seasonal estrus and single birth. The mean litter sizes of Small-tailed Han sheep, Hu sheep, Wadi sheep and Tan sheep have been reported to be 2.67, 2.77, 2.43 and 1.02 (China National Commission of Animal Genetic Resources, 2011), respectively. Based on the essential role of BMPR1B, BMP15 and GDF9 in the regulation of the ovulation rate and terminal folliculogenesis, three fecundity-related genes are considered possible candidate genes for prolificacy traits of sheep. Therefore, the present study was designed with the objectives of polymorphic study of three fecundity-related genes in high-prolificacy breeds (Small-tailed Han sheep, Hu sheep and Wadi sheep) and a low-prolificacy breed (Tan sheep) by direct sequencing after polymerase chain reaction (PCR) amplification.
3.1 Animals and genomic DNA isolation
Blood samples (10 mL, jugular vein, ACD anticoagulant) were collected from 180 Small-tailed Han ewes (Jiaxiang Sheep Breeding Farm, Jiaxiang County, Shandong Province, China), 164 Tan ewes (Tan Sheep Breeding Farm, Yanchi County, Ningxia Hui Autonomous Region, China), 160 Wadi ewes (Zhanhua County, Shandong Province, China) and 172 Hu ewes (Yuhang Hu Sheep Breeding Farm, Hangzhou, Zhejiang Province, China). Genomic DNA was extracted from blood using the phenol-chloroform method, dissolved in TE buffer (10 mmol L−1 Tris-HCl [pH 8.0], 1 mmol L−1 EDTA [pH 8.0]) and kept at −20∘.
3.2 DNA sequencing
According to the DNA sequences of sheep (Ovis aries) (BMPR1B, NC_056059.1; BMP15, NC_056080.1; GDF9, NC_056058.1) and mRNA sequences of sheep (Ovis aries) (BMPR1B, XM_060416665.1; BMP15, NM_001114767.2; GDF9, NM_001142888.2), a total of five pairs of PCR primers were designed using Oligo 6.0 to amplify the sixth exon of BMPR1B (BMPR1B-6), the second exon of BMP15 (BMP15-2) and the first (GDF9-1) and second (GDF9-2-1, GDF9-2-2) exons of GDF9. The primers were synthesized by Shenzhen BGI Co. Ltd. (Shenzhen, China). The primers, predicted product sizes and annealing temperatures are listed in Table 1.
PCR was carried out in a 20 µL volume containing 0.15 µmol L−1 primer, 1× PCR buffer (50 mmol L−1 KCl, 10 mmol L−1 Tris-HCl [pH 8.0], 0.1 % Triton X-100), 2.0 mmol L−1 MgCl2, 0.2 mmol L−1 dNTP, 100 ng ovine genomic DNA and 0.05 U µL−1 Taq DNA polymerase (Promega, Madison, WI, USA), and the rest was ddH2O. The amplification conditions were as follows: initial denaturation at 94∘ for 5 min, followed by 35 cycles of denaturation at 94∘ for 30 s, annealing at 54–55∘ for 45 s, extension at 72∘ for 35 s, and a final extension at 72∘ for 10 min on Mastercycler® 5333 (Eppendorf AG, Hamburg, Germany). The PCR products were directly sequenced using the ABI3730 automatic sequencer (Perkin Elmer Applied Biosystems, Foster City, CA, USA) by Shenzhen BGI Co. Ltd. (Shenzhen, China).
3.3 Statistical analysis
Genotype distributions were analyzed using a chi-squared test. Linkage disequilibrium and haplotype analyses were performed using the SHEsis (Shi and He, 2005; Li et al., 2009) and Phase 2.1.1 (Stephens et al., 2001; Stephens and Donnelly, 2003) software.
In the study, six SNPs were identified in exons of three fertility-related genes (BMPR1B, BMP15 and GDF9) based on DNA sequence genotyping. Two new mutations, c.T755C (S1) in BMP15 and c.A1515G (S2) in GDF9, were identified by analyzing four sheep breeds along with two other known mutations in GDF9 (c.G477A, G3, and c.G721A, G4), and only c.A746G (FecB) in BMPR1B and c.T755C (G1) in BMP15 were detected, in agreement with the 11 mutations in the previous study.
4.1 Genotyping of the BMPR1B mutation
BMPR1B had two alleles: the “B” represented the FecB mutant nucleotide (carrier), and the “+” represented the wild-type nucleotide (non-carrier) (Fig. 1). All three genotypes (, B+ and BB) were observed in the Small-tailed Han sheep, Wadi sheep and Tan sheep, while the Hu sheep lacked the genotype. The FecB mutation changes glutamine to arginine in sheep. In this study, the B allele and BB genotype were predominant in the Hu sheep and Small-tailed Han sheep. In contrast, the + allele and genotype were predominant in the Tan sheep (Table 2). Genotype distributions were significantly different between the four sheep breeds (Table 3).
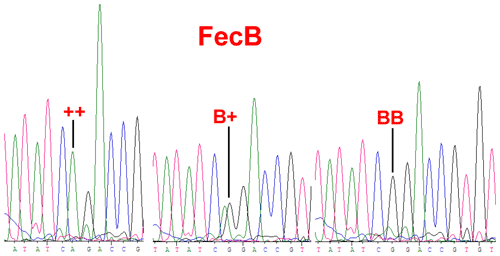
Figure 1Sequence of genotypes , B+ and BB at the 746 loci in the Coding Sequence (CDS) region of BMPR1B sheep.
Table 2Allele and genotype frequencies of the SNPs of the three fecundity genes in the four sheep breeds.
4.2 Genotyping of BMP15 mutations
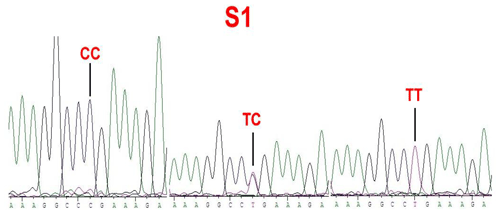
No mutations (FecXI, FecXH, FecXB, FecXG, FecXL and FecXR) of the BMP15 were detected in the four sheep breeds. However, a novel point mutation (T755C) (named S1) in the sheep BMP15 was observed in the four sheep breeds, causing the substitution of a large nonpolar group for a small nonpolar group (Leu252Pro). The S1 mutation had three genotypes, CC, TC and TT (Fig. 2), and the T allele and TT genotype were predominant in the four sheep breeds (Table 2). There were significant differences in the genotype distribution between Wadi sheep and Small-tailed Han sheep (P<0.001), Tan sheep (P<0.01) and Hu sheep (P<0.05), respectively. There was no difference in the genotype distribution among the other three sheep breeds (Table 3).
4.3 Genotyping of GDF9 mutation
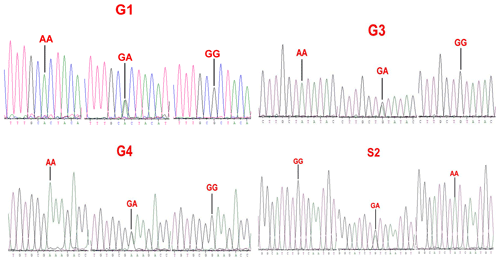
Sequence alignment results showed that four nucleotide mutations were found in the sheep GDF9 gene (Fig. 3), in which a point mutation (G260A) was observed in exon 1 and three SNPs (G477A, G721A and A1515G) were identified in exon 2. Of these four mutations, G260A, G477A and G721A were known as mutations G1, G3 and G4, respectively, which were reported by Hanrahan et al. (2004), and a new point mutation (A1515G) (named S2) was detected in the four sheep breeds. The G3 and S2 mutations were nucleotide changes that did not cause an altered amino acid. G1 and G4 led to amino acid changes, Arg-His and Glu-Lys, respectively. All four mutations gave rise to three genotypes. Small-tailed Han and Wadi sheep lacked AA at the G1 locus. We only detected GG at the S2 locus in Small-tailed Han sheep and AA at the G4 locus in Tan sheep (Table 2).
4.4 The haplotype and diplotype analysis of GDF9
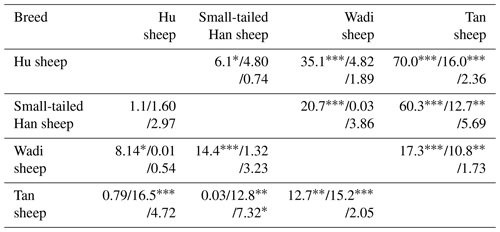
Interestingly, at the G1 and G4 loci, there were significant differences in the genotype distribution between Tan sheep (low prolificacy) and the other three sheep breeds (high prolificacy). There was no difference in the genotype distribution among the other three sheep breeds at the G1 and G4 loci. For the G3 and S2 loci, except for Tan sheep and the Small-tailed Han sheep (P<0.05) at S2, there was no significant difference (P>0.05) in the genotype distribution among the four sheep breeds (Table 3).
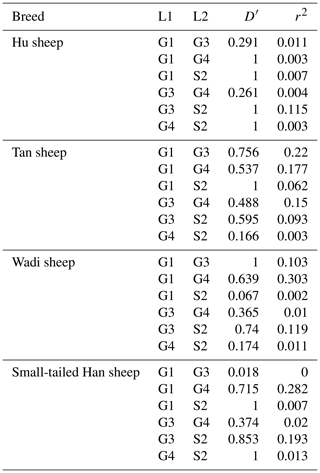
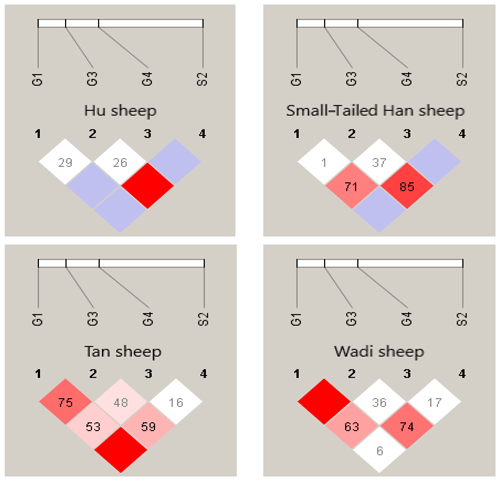
There were 10 haplotypes (Table 4) and 21 diplotypes (Table 5; this was listed when the frequency was larger than 1 %) in the four sheep breeds. H4 was the preponderant haplotype in the three high-prolificacy sheep breeds, while H1 was for Tan sheep. H1 and H4 were the predominant diplotypes in all four sheep breeds. The four sheep breeds were all in linkage equilibrium, and their r2 was less than 0.8. The results are shown in Table 6 and Fig. 4.
Table 4Haplotype frequencies of GDF9 in the four sheep breeds.
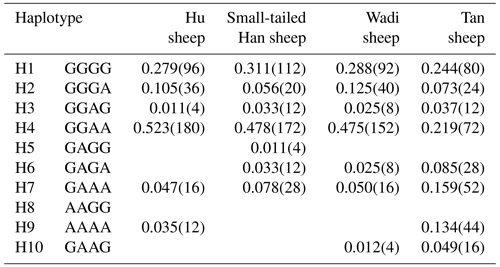
The numbers of haplotypes are in the parentheses.
Table 5Diplotype frequencies of GDF9 in the four sheep breeds.
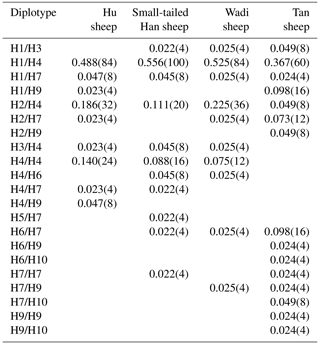
The numbers of diplotypes are in parentheses.
In sheep, the BMPR1B, BMP15 and GDF9 genes were found to be closely associated with a genetic basis for prolificacy (Lv et al., 2022; Ji et al., 2023; Zhu et al., 2023). Abdoli et al. (2013) detected polymorphisms of GDF9 exon 1 and BMPR1B exon 8 in Mehraban ewes. The results showed that two SNPs were detected in both genes, and BMPR1B exon 8 genotypes appear to have significant effects on reproduction traits, while the GDF9 genotypes did not have this role. Furthermore, ewes with mutations in both BMP15 and BMPR1B had greater litter sizes than those with either mutation separately, and the effect of the BMPR1B mutation was greater than that of the BMP15 mutation on litter size in sheep (Davis et al., 1999; Davis, 2005; Chu et al., 2007a). To further investigate the type and frequency of mutations in these three genes in Chinese indigenous breeds, we examined the available SNP loci to complement the polymorphism studies of these three candidate genes associated with prolificacy traits in sheep.
BMPR1B (FecB) was a dominant, autosomal gene that had an additive effect on the ovulation rate and litter size (Davis, 2004). The FecB locus is mapped to sheep chromosome 6, corresponding to human chromosome 4 that contains the BMPR1B. For the FecB mutation, a single A–G substitution at nucleotide position 746 gave rise to an arginine replacing a glutamine (Wilson et al., 2001). Many previous findings identified that the FecB was a major gene for high-prolificacy traits in sheep. The FecB mutation was initially found in Booroola sheep (Mulsant et al., 2001; Souza et al., 2001; Wilson et al., 2001). Subsequent studies showed that the FecB mutation was present in 14 sheep breeds, and half of them were native breeds of China (Chu et al., 2007b; Mahdavi et al., 2014). However, there were some sheep breeds with no FecB mutation, including the Chinese Merino, Tan, Dorset and another 24 sheep breeds (Zuo et al., 2013; Jamshidi et al., 2013). One copy of FecB increased the ovulation rate by 1.3–1.6 and two copies by 2.7–3.0; the litter size is increased by 0.9–1.2 in ewes carrying a single copy and 1.1–1.7 in ewes with two copies (Piper and Bindon, 1982; Bindon, 1984; Piper et al., 1985). In Hu sheep, Small-tailed Han sheep, Wadi sheep and Altay sheep, ewes with genotype BB or B+ had 1.17 (P<0.001) or 1.02 (P<0.001) lambs more than those with genotype (Chu et al., 2011; Li et al., 2012). In this study, the FecB mutation was detected in the four (high- and low-prolificacy) sheep breeds, and the frequency of B allele was significantly different in the high-prolificacy breeds than that in the low-prolificacy breed. This result agreed with the previous studies on Chinese sheep breeds, whose fecundities were similar to those reported in Booroola sheep (Reader et al., 2012).
Another important prolificacy-related gene is BMP15, which is located on the X chromosome, and five mutations and one deletion are known in sheep breeds. These mutations comprise premature stop codons FecXH and FecXG, nonsynonymous amino acid substitution FecXI, FecXB and FecXL and a 17 bp deletion of the reading frame FecXR (Galloway et al., 2000; Hanrahan et al., 2004; Bodin et al., 2007; Martinez-Royo et al., 2008; Monteagudo et al., 2009). The above six mutations in BMP15 were associated with both increased ovulation rates and litter sizes in heterozygous carriers as well as sterility in homozygous carriers (Hanrahan et al., 2004). Studies suggested that heterozygous ewes for these mutations usually increased ovulation rates (without drastic changes in gonadotrophin secretion), while performances of homozygous ewes were early streak ovaries, folliculogenesis arrest and ovarian dysgenesis. The heterozygous deletions reduced BMP15 concentration, increased granulosa cell follicle-stimulating hormone receptor (FSHR) concentration, elevated estrogen secretion and activated the production of stem cell factors (SCFs) (Abir and Fisch, 2011). Without bone morphogenetic protein 15, oocytes will degenerate in the absence of granulosa cell proliferation (Galloway et al., 2000). These findings highlighted the importance of BMP15 in regulating ovulation rates. In our study, no polymorphism was detected in any of these six mutations of sheep BMP15. However, the novel mutation S1 (T755C) was observed in the four sheep breeds, causing the substitution of a large nonpolar group for a small nonpolar group (Leu252Pro). This mutation might be important in causing differences in lambing numbers in native Chinese sheep breeds.
In sheep, GDF9 has been mapped to chromosome 5 and has encoded a prepropeptide of 453 amino acid residues, which could generate an active mature peptide with 135 amino acids (Bodensteiner et al., 1999). GDF9 plays a crucial role as a growth and differentiation factor secreted by oocytes during early folliculogenesis in female reproduction (Elvin et al., 1999). Many findings indicated that GDF9 could regulate several essential granulosa cell enzymes which participated in cumulus expansion and maintenance of an optimal oocyte microenvironment through oocyte–somatic cell interaction and synergistic action (Yan et al., 2001; McNatty et al., 2005b). Several mutations have been observed in sheep GDF9. Li et al. (2003) detected a point mutation (A152G) of the GDF9 gene in Hu, Dorset and Suffolk sheep that gave rise to an amino acid change (Asn51Asp). Eight DNA variants (G1–G8) in the entire GDF9 coding region of Cambridge and Belclare sheep were observed (Hanrahan et al., 2004). FecTT mutation (A1279C) was found in the GDF9 of Icelandic Thoka sheep, leading to a change in serine (neutral) to arginine (basic) at the 109th residue in a mature GDF9 peptide (Nicol et al., 2009). FecGE mutation (T1034G) in the GDF9 gene of Brazilian Santa Ines sheep has been detected (Silva et al., 2011). Chu et al. (2011) found one novel single-nucleotide mutation (G729T) in exon 2 of GDF9, which led to an amino acid change (Gln243His). For all these mutations, heterozygous ewes with the G1, G8 or FecTT mutations have shown an increase in ovulation rate and fecundity. In contrast, the FecGE mutation in GDF9 led to an increased ovulation rate in homozygous ewes (Hanrahan et al., 2004; Nicol et al., 2009; Silva et al., 2011). In this study, four mutations (G1 [G260A], G3 [G477A], G4 [G721A] and S2 [A1515G]) have been observed in the four sheep breeds, of which the S2 mutation did not result in an altered amino acid. Our results were similar to those of previous studies. In particular, we first detected the G1, G3 and G4 mutations simultaneously in Wadi sheep, which are a high-prolificacy local sheep breed in China. Haplotype analysis revealed that H4 (GGAA) was the preponderant haplotype in the three high-prolificacy breeds, but H1 (GGGG) was in Tan sheep (Table 4). H1 and H4 were the predominant diplotypes in all the sheep breeds (Table 5).
In conclusion, we explored polymorphisms in three critical fecundity genes in the Hu sheep, Small-tailed Han sheep, Wadi sheep and Tan sheep breeds. For BMPR1B, the FecB mutation was present in all four sheep breeds. The frequency of B allele in the high-prolificacy breeds was significantly higher than that in the low-prolificacy breed (P<0.001). For BMP15, a novel mutation (c.T755C) was found, while six known mutations (FecXI, FecXH, FecXB, FecXG, FecXL and FecXR) were not detected in the four sheep breeds. For GDF9, the three known mutations (FecGH (G8), FecGE and FecTT) were not detected, and three known loci (G1, G3 and G4) as well as a new mutation, c.A1515G (named S2), were found in the four sheep breeds. The predominant alleles and genotypes are different between the high- and low-prolificacy breeds. At the G1 and G4 loci, there were significant differences in the genotype distribution between Tan sheep (low prolificacy) and the other three sheep breeds (high prolificacy), and there was no difference in the genotype distribution between the other three sheep breeds. Haplotype analysis of the four polymorphic loci in GDF9 indicated that H4 (GGAA) and H1 (GGGG) were the preponderant haplotypes in the high-prolificacy sheep breeds and the low-prolificacy breed, respectively. These results preliminarily showed that the BMPR1B and GDF9 might be major genes influencing the prolificacy of Chinese sheep breeds.
No data sets were used in this article.
Conceptualization: JW and YL. Methodology: WW, YL, SG, RD, XW and XH. Software: JW, YL, SG, RD, XW and XH. Writing (original draft preparation): JW. Writing (review and editing): YL and MC. Supervision: MC. Project administration: MC. Funding acquisition: MC. Reading and agreement to the published version of the paper: all the authors.
The contact author has declared that none of the authors has any competing interests.
All the experiments involving animals were authorized by the Animal Ethics Committee of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (grant no. IAS2020-62).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research was funded by the Xinjiang Uygur Autonomous Region Rural Revitalization Industry Development Science and Technology Action Project (grant no. 2022NC116), the National Natural Science Foundation of China (grant no. 32172704), the Earmarked Fund for China Agriculture Research System of MOF and MARA (grant no. CARS-38) and the Agricultural Science and Technology Innovation Program of China (grant nos. CAAS-ZDRW202106 and ASTIP-IAS13).
This paper was edited by Henry Reyer and reviewed by two anonymous referees.
Abdoli, R., Zamani, P., Deljou, A., and Rezvan, H.: Association of BMPR-1B and GDF9 genes polymorphisms and secondary protein structure changes with reproduction traits in Mehraban ewes, Gene, 524, 296–303, 2013.
Abdoli, R., Zamani, P., Mirhoseini, S. Z., Ghavi, H. Z. N., and Nadri, S.: A review on prolificacy genes in sheep, Reprod. Domest. Anim., 51, 631–637, 2016.
Abir, R. and Fisch, B.: A single nucleotide polymorphism in BMP15 is associated with high response to controlled ovarian hyperstimulation, Reprod. BioMed. Online, 23, 77–80, 2011.
Bindon, B. M.: Biology of the Booroola Merino sheep, Aust. J. Biol. Sci., 37, 163–189, 1984.
Bodensteiner, K. J., Clay, C. M., Moeller, C. L., and Sawyer, H. R.: Molecular cloning of the ovine growth/differentiation factor-9 gene and expression of growth/differentiation factor-9 in ovine and bovine ovaries, Biol. Reprod., 60, 381–386, 1999.
Bodin, L., Di, P. E., Fabre, S., Bontoux, M., Monget, P., Persani, L., and Mulsant, P.: A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep, Endocrinology, 148, 393–400, 2007.
Chu, M., Jia, L., Zhang, Y., Jin, M., Chen, H., Fang, L., Di, R., Cao, G., Feng, T., Tang, Q., Ma, Y., and Li, K.: Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep, Mol. Biol. Rep., 38, 4071–4076, 2011.
Chu, M. X., Liu, Z. H., Jiao, C. L., He, Y. Q., Fang, L., Ye, S. C., Chen, G. H., and Wang, J. Y.: Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries), J. Animal Sci., 85, 598–603, 2007a.
Chu, M. X., Sun, J., Chen, H. Q., and Fang, L.: Detection of the FecXL mutation of BMP15 gene in sheep, Chinese Agricultural Science Bulletin, 23, 85–88, 2007b.
Chu, M. X., Yang, J., Feng, T., Cao, G. L., Fang, L., Di, R., Huang, D. W., Tang, Q. Q., Ma, Y. H., Li, K., and Li, N.: GDF9 as a candidate gene for prolificacy of Small Tail Han sheep, Mol. Biol. Rep., 38, 5199–5204, 2011.
Davis, G. H.: Fecundity genes in sheep, Anim. Reprod. Sci., 82–83, 247–253, 2004.
Davis, G. H.: Major genes affecting ovulation rate in sheep, Genet. Sel. Evol., 37, S11–S23, 2005.
Davis, G. H., Montgomery, G. W., Allison, A. J., Kelly, R. W., and Bray, A. R.: Segregation of a major gene influencing fecundity in progeny of Booroola sheep, New Zeal. J. Agr. Res., 25, 525–529, 1982.
Davis, G. H., Dodds, K. G., and Bruce, G. D.: Combined effect of the Inverdale and Booroola prolificacy genes on ovulation rate in sheep, Proc. Assoc. Adv. Anim. Breed Genet., 13, 1374–1377, 1999.
Demars, J., Fabre, S., Sarry, J., Rossetti, R., Gilbert, H., Persani, L., Tosser-Klopp, G., Mulsant, P., Nowak, Z., Drobik, W., Martyniuk, E., and Bodin, L.: Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep, PLoS Genet., 9, e1003482, https://doi.org/10.1371/journal.pgen.1003482, 2013.
Di, R., Wang, F., Yu, P., Wang, X., He, X., Mwacharo, J. M., Pan, L., and Chu, M.: Detection of Novel Variations Related to Litter Size in BMP15 Gene of Luzhong Mutton Sheep (Ovis aries), Animals (Basel), 11, 3528, https://doi.org/10.3390/ani11123528, 2021.
Elvin, J. A., Clark, A. T., Wang, P., Wolfman, N. M., and Matzuk, M. M.: Paracrine actions of growth differentiation factor-9 in the mammalian ovary, Mol. Endocrinol., 13, 1035–1048, 1999.
Galloway, S. M., McNatty, K. P., Cambridge, L. M., Laitinen, M. P., Juengel, J. L., Jokiranta, T. S., McLaren, R. J., Luiro, K., Dodds, K. G., Montgomery, G. W., Beattie, A. E., Davis, G. H., and Ritvos, O.: Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner, Nat. Genet., 25, 279–283, 2000.
Hanrahan, J. P., Gregan, S. M., Mulsant, P., Mullen, M., Davis, G. H., Powell, R., and Galloway, S. M.: Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries), Biol. Reprod., 70, 900–909, 2004.
Jamshidi, R., Kasirian, M. M., and Rahimi, G. A.: Application of PCR-RFLP technique to determine Booroola gene polymorphism in the Sangsari sheep breed of Iran, Turkish Journal of Veterinary and Animal Sciences, 37, 129–133, 2013.
Ji, X., Cao, Z., Hao, Q., He, M., Cang, M., Yu, H., Ma, Q., Li, X., Bao, S., Wang, J., and Tong, B.: Effects of new mutations in BMPRIB, GDF9, BMP15, LEPR, and B4GALNT2 genes on litter size in sheep, Vet Sci., 10, 37104413, https://doi.org/10.3390/vetsci10040258, 2023.
Li, B. X., Chu, M. X., and Wang, J. Y.: PCR-SSCP analysis on growth differentiation factor 9 gene in sheep, Acta Genet Sinica, 30, 307–310, 2003.
Li, D., Sun, W., Ni, R., Zhang, X. N., Zhang, Y. L., Chu, M. X., Zhang, Y. F., Shen, G. Z., Chen, L., Wu, W. Z., Zhou, H., Han, F., and Xu, H. S.: Genetic diversity of FecB gene and association analysis of its litter size, Journal of Animal Science and Veterinary Medicine, 31, 1–8, 2012.
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., He, L., and Shi, Y.: Apartition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn), Cell Res., 19, 519–523, 2009.
Lv, F. H., Cao, Y. H., Liu, G. J., Luo, L. Y., Lu, R., Liu, M. J., Li, W. R., Zhou, P., Wang, X. H., Shen, M., Gao, L., Yang, J. Q., Yang, H., Yang, Y. L., Liu, C. B., Wan, P. C., Zhang, Y. S., Pi, W. H., Ren, Y. L., Shen, Z. Q., Wang, F., Wang, Y. T., Li, J. Q., Salehian-Dehkordi, H., Hehua, E., Liu, Y. G., Chen, J. F., Wang, J. K., Deng, X. M., Esmailizadeh, A., Dehghani-Qanatqestani, M., Charati, H., Nosrati, M., Štěpánek, O., Rushdi, H. E., Olsaker, I., Curik, I., Gorkhali, N. A., Paiva, S. R., Caetano, A. R., Ciani, E., Amills, M., Weimann, C., Erhardt, G., Amane, A., Mwacharo, J. M., Han, J. L., Hanotte, O., Periasamy, K., Johansson, A. M., Hallsson, J. H., Kantanen, J., Coltman, D. W., Bruford, M. W., Lenstra, J. A., and Li, M. H.: Whole-genome resequencing of worldwide wild and domestic sheep elucidates genetic diversity, introgression, and agronomically important loci, Mol. Biol. Evol., 39, msab353, https://doi.org/10.1093/molbev/msab353, 2022.
Mahdavi, M., Nanekarani, S., and Hosseini, S. D.: Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep, Anim. Reprod. Sci., 147, 93–98, 2014.
Martinez-Royo, A., Jurado, J. J., Smulders, J. P., Martí, J. I., Alabart, J. L., Roche, A., Fantova, E., Bodin, L., Mulsant, P., Serrano, M., Folch, J., and Calvo, J. H.: A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep, Anim. Genet., 39, 294–297, 2008.
McNatty, K. P., Juengel, J. L., Reader, K. L., Lun, S., Myllymaa, S., Lawrence, S. B., Western, A., Meerasahib, M. F., Mottershead, D. G., Groome, N. P., Ritvos, O., and Laitinen, M. P.: Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function in ruminants, Reproduction, 129, 481–487, 2005a.
McNatty, K. P., Smith, P., Moore, L. G., Reader, K., Lun, S., Hanrahan, J. P., Groome, N. P., Laitinen, M., Ritvos, O., and Juengel, J. L.: Oocyte-expressed genes affecting ovulation rate, Mol. Cell. Endocrinol., 234, 57–66, 2005b.
Melo, E. O., Silva, B. D., Castro, E. A., Silva, T., Paiva, S. R., Sartori, R., Franco, M. M., Souza, C. J., and Neves, J. P.: A novel mutation in the growth and differentiation factor 9 (GDF9) gene is associated, in homozygosis, with increased ovulation rate in Santa Ines sheep, Biol. Reprod., 78, 371, https://doi.org/10.1093/biolreprod/78.s1.141b, 2008.
Monteagudo, L. V., Ponz, R., Tejedor, M. T., Lavi, A., and Sierra, I.: A 17 bp deletion in the bone morphogenetic protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed, Anim. Reprod. Sci., 110, 139–146, 2009.
Mulsant, P., Lecerf, F., Fabre, S., Schibler, L., Monget, P., Lanneluc, I., Pisselet, C., Riquet, J., Monniaux, D., Callebaut, I., Cribiu, E., Thimonier, J., Teyssier, J., Bodin, L., Cognie, Y., and Elsen, J. M.: Mutation in bone morphogenetic protein receptor-1B is associated with increased ovulation rate in Booroola Merino ewes, P. Natl. Acad. Sci. USA, 98, 5104–5109, 2001.
Nicol, L., Bishop, S. C., Pong-Wong, R., Bendixen, C., Holm, L. E., Rhind, S. M., and McNeilly, A. S.: Homozygosity for a single base-pair mutation in the oocyte-specific GDF9 gene results in sterility in Thoka sheep, Reproduction, 138, 921–933, 2009.
Piper, L. R. and Bindon, B. M.: The Booroola Merino and the performance of medium non-Peppin crosses at Armidale, in: The Booroola Merino, edited by: Piper, L. R., Bindon, B. M., and Nethery, R. D., CSIRO, Melbourne, Australia, ISBN 0643030239, 1982.
Piper, L. R., Bindon, B. M., and Davis, G. H.: The single gene inheritance of the high litter size of the Booroola Merino, in: Genetics of Reproduction in Sheep, edited by: Land, R. B. and Robinson, D. W., Butterworths, London, UK, ISBN 9780407003026, 1985.
Reader, K. L., Haydon, L. J., Littlejohn, R. P., Juengel, J. L., and McNatty, K. P.: Booroola BMPR1B mutation alters early follicular development and oocyte ultrastructure in sheep, Reprod. Fert. Develop., 24, 353–361, 2012.
Shi, Y. Y. and He, L.: SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci, Cell Res., 15, 97–98, 2005.
Silva, B. D., Castro, E. A., Souza, C. J., Paiva, S. R., Sartori, R., Franco, M. M., Azevedo, H. C., Silva, T. A., Vieira, A. M., Neves, J. P., and Melo, E. O.: A new polymorphism in the Growth and Differentiation Factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep, Anim. Genet., 42, 89–92, 2011.
Souza, C. J., MacDougall, C., Campbell, B. K., McNeilly, A. S., and Baird, D. T.: The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1B (BMPR1B) gene, J. Endocrinol., 169, R1–R6, 2001.
Stephens, M. and Donnelly, P.: A comparison of Bayesian methods for haplotype reconstruction from population genotype data, Am. J. Hum. Genet., 73, 1162–1169, 2003.
Stephens, M., Smith, N. J., and Donnelly, P.: A new statistical method for haplotype reconstruction from population data, Am. J. Hum. Genet., 68, 978–989, 2001.
Stocker, W. A., Walton, K. L., Richani, D., Chan, K. L., Beilby, K. H., Finger, B. J., Green, M. P., Gilchrist, R. B., and Harrison, C. A.: A variant of human growth differentiation factor-9 that improves oocyte developmental competence, J. Biol. Chem., 295, 7981–7991, 2020.
Wang, F., Chu, M., Pan, L., Wang, X., He, X., Zhang, R., Tao, L., La, Y., Ma, L., and Di, R.: Polymorphism detection of GDF9 gene and its association with litter size in Luzhong mutton sheep (Ovis aries), Animals (Basel), 11, 571, https://doi.org/10.3390/ani11020571, 2021.
Wen, Y.-L., Guo, X.-F., Ma, L., Zhang, X.-S., Zhang, J.-L., Zhao, S.-G., and Chu, M.-X.: The expression and mutation of BMPR1B and its association with litter size in small-tail Han sheep (Ovis aries), Arch. Anim. Breed., 64, 211–221, https://doi.org/10.5194/aab-64-211-2021, 2021.
Wilson, T., Wu, X. Y., Juengel, J. L., Ross, I. K., Lumsden, J. M., Lord, E. A., Dodds, K. G., Walling, G. A., McEwan, J. C., O'Connell, A. R., McNatty, K. P., and Montgomery, G. W.: Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells, Biol. Reprod., 64, 1225–1235, 2001.
Yan, C., Wang, P., DeMayo, J., DeMayo, F. J., Elvin, J. A., Carino, C., Prasad, S. V., Skinner, S. S., Dunbar, B. S., Dube, J. L., Celeste, A. J., and Matzuk, M. M.: Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function, Mol. Endocrinol., 15, 854–866, 2001.
Zhu, M., Yang, Y., Yang, H., Zhao, Z., Zhang, H., Blair, H. T., Zheng, W., Wang, M., Fang, C., Yu, Q., Zhou, H., and Qi, H.: Whole-genome resequencing of the native sheep provides insights into the microevolution and identifies genes associated with reproduction traits, BMC Genomics, 24, 392, https://doi.org/10.1186/s12864-023-09479-y, 2023.
Zuo, B., Qian, H., Wang, Z., Wang, X., Nisa, N., Bayier, A., and Wang, F.: A study on BMPR-IB gene of Bayanbulak sheep, Asian Austral. J. Anim. Sci., 26, 36–42, 2013.