the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
A comprehensive analysis of the effects of DGAT1 K232A polymorphism on milk production and fertility traits in Holstein Friesian and Jersey cows reared in Türkiye
Sena Ardicli
Ertugrul Kul
Samet Hasan Abaci
Eser Kemal Gurcan
Soner Cankaya
Research on the diacylglycerol acyltransferase 1 (DGAT1) K232A marker in cattle shows inconsistent results across regions, largely due to small sample sizes, limited genetic variation, and data restricted to few lactations, which complicates establishing a reliable genotype–phenotype correlation. This research aimed to determine the effect of the K232A polymorphism of the bovine DGAT1 gene on milk production and quality traits in dairy cattle. We used 1104 cattle, including 828 Holstein Friesian and 276 Jersey cows. The analysis utilized extensive data from six lactations of cows raised on four commercial dairy farms. We genotyped the population using the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) technique and Sanger sequencing for verification. We then evaluated the 305 d and test-day milk yields as well as fat and protein yields and percentages. The number of inseminations per conception and calving ease were also assessed as reproduction indices. Genotype–phenotype associations were quantified using linear mixed models. The AA genotype was absent in Jersey cows, and the heterozygous genotype was predominant in both breeds. The K232A marker was significantly associated with test-day milk yield, fat, and protein content in Jersey cows. Further, it substantially affected the fat percentage of milk in Holstein Friesian cows (p<0.001). We found that the KK genotype is highly desirable for milk quality and especially fat content. This comprehensive assessment demonstrated that the KK genotype of the DGAT1 K232A polymorphism significantly influenced fat and protein contents in dairy cattle.
- Article
(2060 KB) - Full-text XML
-
Supplement
(408 KB) - BibTeX
- EndNote
Milk production is a phenotypic trait determined by the genotype and the environment that shows a large degree of variability in dairy cows. Many genes with additive effects determine the quantity and quality of milk; thus, these traits follow a polygenic inheritance model. By choosing the genes thought to be related to milk production and the appropriate genotypes of these genes, it will be possible to make more accurate predictions of genetic values for selection candidates and speed up the selection process.
Several researchers have focused on the BTA14 regarding the connection between particular quantitative trait loci (QTLs) and milk yield/composition traits – primarily fat content (Ashwell et al., 2004; Fink et al., 2020; Weller et al., 2003). Thus, many candidate genes associated with bovine milk production traits have been identified in recent decades (Fig. 1a). One of the most famous genes located on BTA14 is the diacylglycerol acyltransferase 1 (DGAT1) gene (GenBank accession number AJ318490). This gene significantly affects metabolism and especially intestinal fat absorption, adipose tissue formation, and lipoprotein assembly (Mohammed et al., 2015). The bovine DGAT1 encodes an enzyme with the same name (DGAT1), which catalyzes the final step in triglyceride synthesis (Kong et al., 2007; Thaller et al., 2003). This multipass transmembrane protein acts as a crucial metabolic enzyme, converting diacylglycerol and fatty acyl coenzyme A to triacylglycerol (Lehner and Kuksis, 1996). The gene DGAT1, responsible for encoding this biochemically important enzyme, is expressed in almost all tissues, including mammary glands (Mahmoudi and Rashidi, 2023). It is on the centromeric region of BTA14 (Grisart et al., 2002; Mahmoudi and Rashidi, 2023; Winter et al., 2002), located in the 603813-612791 forward strand, and consists of 17 exons and 16 introns (Ensembl, 2024). Several studies reported a strong association between the bovine DGAT1 and various milk-related traits in dairy cattle (Anton et al., 2012; Ardicli et al., 2018; Bobbo et al., 2018; Gothwal et al., 2022; Grisart et al., 2002, 2004; Winter et al., 2002).
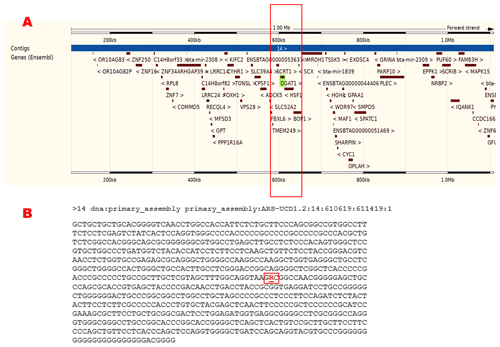
Figure 1Brief information about the region in detail for the bovine diacylglycerol O-acyltransferase 1 (DGAT1), primary_assembly 14:603813–612791 (forward strand; ARS-UCD1.2:CM008181.2). (a) The genomic location, that is, the nearest genes of the DGAT1 gene in the cattle genome. (b) The flanking sequence represents the DGAT1 K232A variant (rs109234250). The figure was created using the Ensembl genome browser (2024; https://www.ensembl.org/Bos_taurus/Gene/Summary?db=core;g=ENSBTAG00000026356;r=14:603035-612781, last access: 27 September 2024).
Concerning the bovine DGAT1 gene sequence, the dinucleotide change (AA/GC) at positions 10433 (g.10433G>A) and 10434 (g.10434C>A) in exon 8 leads to a non-conservative substitution of lysine by alanine at amino acid position 232 (Gothwal et al., 2022). Increased milk content in different breeds is strongly associated with the lysine position 232 of the protein encoded by bovine DGAT1; an alanine at this position is associated with lower milk fat content (Winter et al., 2002). The origin of this causal mutation (Mahmoudi and Rashidi, 2023) can be traced back to cattle domestication (Grisart et al., 2002; Winter et al., 2002). The lysine encoding variant is the ancestral constituent of DGAT1. The K232A alteration existed in the early stages of, or even before, bovine domestication (Winter et al., 2002). The presence of an alanine variant in the indigenous Anatolian Black cattle breed points out clues for the history of this alteration because these cattle have been living at the site of the domestication of European Bos taurus (Medjugorac et al., 1994). The subspecies Bos taurus indicus, which was domesticated separately, does not carry the alanine variant (Loftus et al., 1994). This mutation plays a critical role in triacylglycerol metabolism by affecting the activity of the resultant enzyme (Grisart et al., 2002; Mahmoudi and Rashidi, 2023; Winter et al., 2002). Many researchers have demonstrated that DGAT1 K232A polymorphism has impacts on milk yield (Anton et al., 2008; Ardicli et al., 2018; Bovenhuis et al., 2016; Gothwal et al., 2022; Li et al., 2021; Manga et al., 2011; Mao et al., 2012; Samuel et al., 2023; Schennink et al., 2007; Vanbergue et al., 2016), fat content (Carvajal et al., 2016; Li et al., 2021; Mahmoudi and Rashidi, 2023; Manga et al., 2011; Mao et al., 2012), and protein content (Anton et al., 2008; Li et al., 2021; Mahmoudi and Rashidi, 2023; Mao et al., 2012; Vanbergue et al., 2016). On the other hand, several studies reported contrary results on milk-related traits (Akyuz et al., 2015; Ardicli et al., 2018; Khan et al., 2019; Krovvidi et al., 2021; Manga et al., 2011; Strzałkowska et al., 2005; Tumino et al., 2021; Valenti et al., 2019; Van Gastelen et al., 2017).
Although the DGAT1 K232A marker has been widely studied in various cattle breeds, the results from different geographical regions often conflict even for the same traits. In many studies, it is striking that some limiting factors prevent the establishment of a reliable genotype–phenotype balance. First, several studies have been conducted on limited sample sizes. Moreover, genotyping dairy populations with insufficient sires results in low genetic variation. The absence of particular genotypes causes ignoring these variants' critical effects. Predominantly, a vast majority of the studies consist of data limited to a few lactations. On the other hand, some essential environmental impacts, such as lactation curve constituents or calving season, are lacking in the statistical analysis. Therefore, we aimed to quantify the effects of the DGAT1 K232A polymorphism (rs109234250) on milk production traits in Holstein Friesian and Jersey dairy cows.
2.1 Animal material and phenotypic records
Animals used in the present study (n=1104) were purebred Jersey (n=276) and Holstein Friesian cows (n=828). All of the Jersey cows were from one and Holstein Friesians were from three commercial dairy herds in the south Marmara region of Türkiye. All animals were raised in free-stall barns and milked two (for Jerseys) and three times (for Holstein Friesians) a day. Cows were milked in parlors fitted with electronic systems that automatically recorded the milk volume. Thus, the milk yield for each cow during each milking session was tracked using DeLaval's Alpro herd management system (DeLaval International, Tumba, Sweden). The mean results of each cow from two (for Jerseys) and three (for Holstein Friesians) consecutive milkings were used. Milk samples were taken individually on milk test days monthly for six lactations. Then, 305 d milk yield (305-DMY) was calculated using the individual daily milk yield record dataset. Thus, we acquired the test-day milk yield (TDMY) and 305-DMY records. From the monthly milk control samples, we obtained data on milk composition traits, including fat content (FC), protein content (PC), test-day fat yield (TDFY), test-day protein yield (TDPY), 305 d fat yield (305-DFY), and 305 d protein yield (305-DPY). Milk composition analysis was evaluated by infrared analysis with a Bentley 2000 mid-infrared instrument (Bentley Instruments, Chaska, MN, USA). The data were obtained from three different herds for the Holstein Friesian breed, whereas the data on Jerseys originate from a single farm. We obtained the number of inseminations per conception (NI) and calving ease records (CE) from herd 3 (n=361). CE was evaluated on a scale according to the following criteria: (1) unassisted calving, (2) calving requiring remote assistance, (3) calving with help, and (4) caesareans applied. Since there were low numbers of animals with category 4, we combined categories 3 and 4 and coded them both as 3 to minimize the severe category problem, as González-Recio et al. (2007) suggested. Regarding the entire population, we gathered lactation rank, calving season, calving year, and days in lactation records for the subsequent quantitative analysis. We excluded the records with missing sire identification and incorrect calving dates from the study. This study complied with the relevant national regulations and institutional policies for the care and use of animals (approval number 2010/6-05).
2.2 DNA extraction and genotyping
We isolated the genomic DNA from whole blood samples (∼ 10 mL) taken into K3EDTA tubes (Vacutest Kima SRL, Italy) from the vena jugularis of the animals. All blood samples were conserved at −20 °C until DNA extraction. We then obtained the purified DNA according to the standard phenol–chloroform–isoamyl alcohol () method as Green (2012) suggested. DNA concentration (ng mL−1) and absorbance ratio ( value) were measured using a NanoDrop spectrophotometer (NanoDrop 2000c, Thermo Fisher Scientific, Wilmington, DE, USA).
The genotyping of DGAT1 K232A polymorphism in exon 8 (Fig. 1b) was performed by the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method and Sanger sequencing. A touch-down PCR protocol was applied with the bovine DGAT1 primer sequences to amplify a 411 bp amplicon, as shown in Table 1. PCR mixture consisted of 50 µM primer (1 µL from each forward and reverse primer), 5 µL (25mM) MgSO4, 1 µL dNTP (200 µM), 5 µL buffer (10x), 1 µL Taq DNA polymerase enzyme, and 33.5 µL ddH2O. We used ∼ 2.50 mL (70–95 ng) genomic DNA for the amplification. Consumables were obtained from Biomatik (Biomatik, Cambridge, Canada) and Thermo (Thermo Fisher Scientific, Inc., Waltham, MA, USA). After PCR, products were electrophoresed using 2 % agarose gels (Sigma Aldrich, Steinheim, Germany) in Tris–Borate–EDTA (TBE) (1×).
Table 1Primers sequences (from 5′ to 3′) and restriction enzyme used for genotyping the DGAT1 locus in the current study.

1 Primer sequences were used as suggested by Winter et al. (2002). 2 A touch-down PCR protocol was used to amplify the 411 bp PCR product. DGAT1: diacylglycerol acyltransferase 1.
In total, 15 µL of PCR product were digested with 0.50–1 mL of the EaeI restriction enzyme (New England Biolabs (NEB), Ipswich, MA, USA, or Thermo Fisher Scientific). We prepared the enzyme–buffer mix consisting of 1 µL restriction enzyme with 8 µL of the respective buffer and 10 µL of the PCR product. The incubation was modified from 3 h to overnight. Amplifications and enzyme incubations were run on two thermal cyclers (Palm Cycler GC1-96, Corbett Research, Sydney, Australia, and T100 Thermal Cycler, Bio-Rad, USA). The digested fragments were separated through 3 % agarose gel electrophoresis and visualized under an automatic gel documentation system (DNR MiniLumi Bio-Imaging Systems, Israel). The predicted bands for each genotype are listed in Table 1.
For further confirmation, we sequenced representative samples from each genotype (n=21) for the bovine DGAT1 gene (GenBank accession number AY065621, genomic sequence NC_037341.1, chromosome 14, reference ARS-UCD2.0 primary assembly). To amplify the target, PCR was performed as mentioned above. The amplified PCR amplicons were purified using the DNA Clean and Concentrator kit (Zymo Research, Orange, California). We sequenced the products following the manufacturer's instructions using the SeqStudio™ Genetic Analyzer with SeqStudio™ Data Collection Software automatic sequencer (Applied Biosystems, Carlsbad, CA). Ultimately, the results were compared and confirmed by the cattle genome provided by the Ensembl genome browser (https://www.ensembl.org/Bos_taurus/Gene/Summary?db=core;g=ENSBTAG00000026356;r=14:603813-612791;t=ENSBTAT00000037423, last access: 27 September 2024).
2.3 Statistical analysis
Allele and genotype frequencies of the DGAT1 K232A polymorphism were calculated based on the suggestions by Falconer and Mackay (1996). The appropriateness to the Hardy–Weinberg equilibrium (HWE) was determined with chi-square (χ2) of observed and predicted genotypic frequencies. If the genotype count was below five, we used Fisher's exact test to evaluate the HWE. Population genetic parameters, including heterozygosity (He), effective allele numbers (Ne), and polymorphism information content (PIC), were calculated according to the formulas stated by Nei and Roychoudhury (1974) and Botstein et al. (1980). We used linear mixed models, selected based on the coefficient of determination (R2) values, in the analysis of variance (ANOVA) to assess the genotype–phenotype association. Minitab (PA, USA, v17.1.0) statistical software was used for the association analysis. The statistical model was as follows:
where Yijklmnop is the phenotypic record, μ is the overall mean, Ai is the fixed effect of the ith calving year (i=2010, 2011, 2012, 2013, 2014, 2015), Bj is the fixed effect of the jth calving season (j is autumn, winter, spring, and summer), Ck is the fixed effect of kth lactation rank (k=1, 2, 3, 4, 5, and 6), Dl is the fixed effect of the lth DGAT1 genotype (l is KK, KA, and AA), β Em is the regression effect of days in lactation, Fn is the impact of animal factor (n is the varying performance value of an individual cow in different lactations), Go is the fixed effect of the oth herd (o=1, 2, and 3), and eijklmnop is the random error.
Initially, the sire effect was included in the model. However, it was excluded because of the minimal number of genotyped bulls that were progenies of the same sire, as Curi et al. (2006) indicated. The model included the herd effect because of three different herds for the Holstein Friesian breed (the data on jerseys originate from a single farm). Furthermore, the model consists of only two genotypes to evaluate the NI and CE since the AA genotype was absent in herd 3. Genotype–environment interactions were either insignificant or, in some cases, meaningless for interpreting the genotype-driven interaction due to the overwhelming environmental influence (e.g., the genotype × lactation rank). Thus, they were not considered further. We analyzed genotypes and milk production traits using least-squares means (LSMs) and their corresponding standard errors (SEs). When significant associations were observed, differences between groups were analyzed using Tukey's post hoc test.
We estimated additive and dominance effects based on a reparameterized model as demonstrated by Falconer and Mackay (1996). In this respect, the additive effect is the difference between the means of two homozygous divided by 2, while the dominance effect is the deviation of the heterozygote from the mean of the two homozygotes.
3.1 The identification of the single-nucleotide polymorphism (SNP)through genotyping
In genotyping the K232A substitution in the bovine DGAT1 gene (primary_assembly 14:611019 (forward strand)) using EaeI restriction enzyme, the approach targets the combined dinucleotide change at positions 10433 and 10434 from AA to GC, which corresponds to the amino acid substitution from lysine to alanine at position 232. The restriction enzyme recognizes a specific sequence, cutting only if this sequence matches the GC variant, thereby indicating the presence of alanine, while the AA sequence, indicative of lysine, remains uncut. In this context, the 411 bp PCR product (Fig. 2) harboring K232A polymorphism of the bovine DGAT1 gene was discriminated by the EaeI restriction enzyme (Y/GGCCR) digestion. The distinct banding characterized the KK genotype by the single undigested band (411 bp). The three-band pattern (411, 208, and 203 bp) was diagnostic for the heterozygote genotype. AA is distinguishable by two bands (208 and 203 bp). Notably, these two bands appear to be a single band (208 + 203 bp) because they are very close (Fig. 3a). However, this does not pose any difficulties in diagnosis. The three genotypes (KK, KA, and AA) were observed in Holstein Friesian cows, but AA was absent in Jerseys (Fig. 3). Following PCR-RFLP, we sequenced the PCR amplicons with a genetic analyzer. The chromatograph representing respective genotypes is shown in Fig. 3b–d. Consequently, the dinucleotide transition from AA to GC at positions 10433 and 10434 (AJ318490.1) in exon 8, which results in a non-conservative amino acid substitution, replacing lysine with alanine at position 232, was properly analyzed.
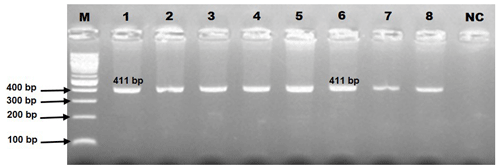
Figure 2The electrophoresis pattern of K232A polymorphism (rs109234250) within the bovine DGAT1 concerning PCR amplification (amplicon size of 411 bp). NC: negative control.
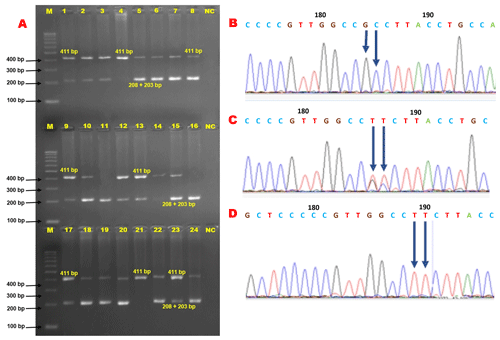
Figure 3Results of PCR-RFLP and Sanger sequencing for bovine DGAT1 K232A polymorphism. (a) The electrophoresis pattern of restriction enzyme digestion of PCR product with EaeI for K232A polymorphism within the bovine DGAT1 gene. Lines 4 and 21: KK; lines 1–3, 5–10, 12–15, 17–20, and 22–24: KA; lines 11 and 16: AA; NC: negative control. Considering the AA genotype, the diagnostic bands (208 and 203 bp) appear to be a single band (208 + 203 bp) because of their closeness. (b) Chromatograph of the K232A locus of the DGAT1 gene representing the AA homozygous genotype. (c) Chromatograph of the K232A locus of the DGAT1 gene representing the heterozygous genotype. (d) Chromatograph of the K232A locus of the DGAT1 gene representing the KK homozygous genotype. The dinucleotide change (AA/GC) at positions 10433 and 10434 in exon 8 leads to a non-conservative substitution of lysine by alanine at position 232. The two alleles at this locus are AA and GC, encoding lysine (K) and alanine (A) at the amino acid position 232. These two alterations in the DGAT1 gene lie immediately adjacent to each another.
3.2 Genetic diversity parameter estimates
Table 2 shows the genotypic distribution, population genetic parameters, and HWE test results. The results are presented separately for both breeds and the whole population. Among 1104 cows, there were only seven animals with the AA genotype. This genotype was absent in the Jersey breed. The frequency of heterozygous animals was predominant in both breeds.
On the other hand, Jersey cows exhibited the highest frequency of the K allele (0.59). We observed concordance with HWE in neither a specific breed nor the entire population. He and Ne values were higher in Holsteins. We also observed the highest PIC in this breed, as shown in Table 2. For both breeds, the marker showed mild variability and informativeness.
Table 2Genotype and allele frequencies, Hardy–Weinberg equilibrium (HWE) test results, and population genetics parameters of the bovine DGAT1 K232A polymorphism in the Holstein Friesian (n=828) and Jersey (n=276) breeds and in the entire dairy population (n=1104).
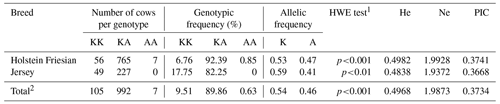
1 Fisher's exact test was used to evaluate the HWE because of the low number of individuals (Jerseys) per genotype (genotype counts below 5). p<0.01; p<0.001 – not consistent with the HWE. 2 Frequencies and parameters were calculated based on the total number of animals (n=1104).
3.3 The association analysis
Table 3 shows the results of the association between the DGAT1 K232A genotypes and milk production traits in the Jersey breed. The present analysis indicated that the cows carrying the KA genotype had a higher TDMY than those with the KK genotype. Moreover, cows carrying the KK genotype showed a higher FC and PC than those with the KA genotype in Jersey cows (p<0.05). It is important to note that there were no Jersey cows with the AA genotype in this study (Table 3). Concerning the Holstein Friesian breed, the effect of the DGAT1 genotypes was statistically significant only for the FC (p<0.001). The KK genotype exhibited remarkably higher FC means compared to AA and heterozygotes. Although this genotype showed a lower TDMY and 305-DMY, the LSM analysis did not substantiate this association at the p<0.05 level (Table 4). Nevertheless, the effect of this genotype can be accepted as a tendency (p<0.1). In herd 3, we evaluated the association between the reproduction traits (NI and CE), but the statistical analysis showed no significance in Holsteins (Table S1).
As shown in Table 5, the dominance effect (d) value of the DGAT1 K232A for TDMY (p<0.01), FC (p<0.001), and PC (p<0.005) was significant in Jersey cows. However, allele substitution effect (a) values were not significant (p>0.05). Concerning the Holstein Friesian breed, a highly substantial additive effect (a) was observed for FC (p<0.001). Dominance effect (d) on all milk production traits was insignificant (p>0.05). On the other hand, a significant allele substitution effect (α) for FC (p<0.001) and protein yield (PY; p<0.05) was detected in Holstein Friesian cows (Table 6).
Table 3Levels of significance, least-squares means, and standard errors for the effect of DGAT1 K232A genotypes on milk production traits in Jersey cows (n=276).

a,b Different letters in a column indicate a significant difference at the p<0.05. TDMY: test-day milk yield, 305-DMY: 305 d milk yield, FC: milk fat content, TDFY: test-day fat yield, PC: milk protein content, TDPY: test-day protein yield, 305-DFY: 305 d fat yield, 305-DPY: 305 d protein yield. 1 TDFY and TDPY (kg d−1) were calculated based on TDMY. 2 p<0.1; a suggestive association (tendency).
Fat and protein are essential milk components and are one of the main reasons for the nutritious property of milk. Milk fats are primarily made up of triglycerides, including one molecule of glycerin and three fatty acids. They are remarkable indicators of milk quality (Pathak et al., 2022). Depending on the fat level, milk has high concentrations of vitamins A and D (Magan et al., 2021). The nutritional value of milk and dairy products is closely associated with the fat content of milk, making up approximately 30 % of the total fat consumed in the human diet (Mansbridge and Blake, 1997; Pathak et al., 2022). Considering cattle milk is the most consumed milk type worldwide, molecular studies on cattle milk composition traits reveal crucial knowledge about human health and nutrition, primarily because of some fatty acids such as phospholipids (Pathak et al., 2022). Many researchers pointed out the DGAT1 as a critical candidate and functional gene for fat traits and milk production (Gothwal et al., 2022; Grisart et al., 2002; Mahmoudi and Rashidi, 2023; Pathak et al., 2022). Several nucleotide alterations in this eminent gene have been associated with critical phenotypic traits in different breeds. Among them, K232A polymorphism (rs109234250) is one of dairy cattle's most famous genetic markers. However, the results of previously published reports are often conflicting. Moreover, there are some conspicuous limitations in these reports, such as limited sample sizes, lack of some genotypes (especially AA), and ignoring some crucial environmental factors in statistical models. In this report, we thoroughly analyze the effects of DGAT1 K232A polymorphism on milk yield and quality in a sizeable population of commercial dairy cattle.
Table 4Levels of significance, least-squares means, and standard errors for the effect of DGAT1 K232A genotypes on milk production traits in Holstein Friesian cows (n=828).
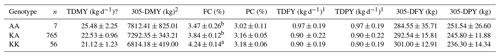
a,b Different superscripts within a column indicate significant differences at the p<0.001 level. TDMY: test-day milk yield, 305-DMY: 305 d milk yield, FC: milk fat content, TDFY: test-day fat yield, PC: milk protein content, TDPY: test-day protein yield, 305-DFY: 305 d fat yield, 305-DPY: 305 d protein yield. 1 TDFY and TDPY (kg d−1) were calculated based on TDMY. 2 p<0.1; a suggestive association (tendency).
Concerning Jersey cows, the effects of DGAT1 genotypes on TDMY (p<0.01), FC (p<0.001), and PC (p<0.01) were statistically significant (Table 3). The cows with the KA genotype had more favorable values for TDMY (+1.11 kg d−1) than cows with the KK genotype. We observed that the KK genotype of the DGAT1 gene was related to higher FC (+0.45 %) and PC (+0.09 %) compared to the KA genotype. Although there were evident differences in the average 305-DMY, fat yield (FY), and PY, they were insignificant (p>0.05). In this study, no Jersey cows with the AA genotype were found (Table 2). Anton et al. (2012) found that the AA-genotype animals had higher values of FC and PC than the other genotypes (5.83 % and 3.96 %, respectively) in Jersey cows. However, these animals exhibited low values for 305-DMY. Bovenhuis et al. (2016) reported that significant effects on milk protein composition in Dutch Holstein Friesians could not be confirmed in Danish Jersey or Danish Holstein Friesian breeds. Komisarek et al. (2004) found that the K allele was associated with high milk fat yield and fat and protein content, whereas the A allele led to increased milk yield in Jersey cows. It is important to note that we observed a lower frequency of the A allele in Jersey cattle. This finding is consistent with earlier reports (Kaupe et al., 2004; Winter et al., 2002; Krovvidi et al., 2021), which indicate that a high frequency of the DGAT1 K allele is a characteristic feature of Jersey cattle. Notably, Ripoli et al. (2006) demonstrated that breeds involved in different lineages affect the genotypic distributions. They stated that European Bos taurus breeds, except the Jersey breed, showed the lowest frequency of the K allele, while cattle of the Bos indicus type harbored the highest K allele frequencies. This situation is one of the most significant reasons for the contradictory results from different breeds regarding milk production traits. It is well known that gene frequencies may differ among different breeds or even different populations of the same breed (Ardicli et al., 2019a). Hence, genotypic results from sizeable dairy populations may reveal more reliable results on genotype–phenotype associations. Several studies have reported that the DGAT1 K232A marker influences milk yield and composition at the same time. Here, we note that this polymorphism impacted the FC and PC but not 305-DMY. Furthermore, the statistically significant effect of the DGAT1 marker turned out into a tendency (p<0.1) only for the fat yield when we expanded the analysis to 305 d lactation (for six lactations). This result again shows how comprehensive data are a vital prerequisite for reliably evaluating the association between single-nucleotide polymorphisms (SNPs) and the milk production traits in commercial dairy herds.
Table 5Genetic effects of DGAT1 K232A polymorphism on milk yield and milk component traits in Jersey breed.
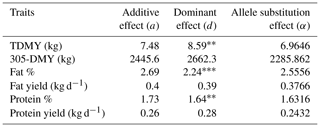
TDMY: test-day milk yield; 305-DMY: 305 d milk yield. Fat and protein yields (kg) were calculated based on TDMY. p<0.01. p<0.001.
DGAT1 is an efficient and functional gene in which alterations may affect several essential traits in dairy cattle. Many authors stated that DGAT1 K232A polymorphism influenced milk yield (Anton et al., 2008; Bovenhuis et al., 2016; Li et al., 2021; Manga et al., 2011; Mao et al., 2012; Molee et al., 2015; Schennink et al., 2007; Sun et al., 2009; Vanbergue et al., 2016). We found no significant association between the DGAT1 marker and milk yield in either of the dairy breeds. Similar results for FC and differences for 305-DMY and PC in Holstein cows were also observed by Anton et al. (2012). Lešková et al. (2013) reported that genotype significantly affected PC but not for 305-DMY and FC in Holstein cows. Contrary to some papers in the literature, we found that the AA-genotype cows tended (p<0.1) to exhibit higher 305-DMY than those with alternative genotypes (Table 4). However, this difference was not corroborated by the statistical analysis. It is important to note that we observed only seven animals carrying this genotype. In general, the frequency of this genotype is remarkably low or absent in Holstein Friesians (Ardicli et al., 2018, 2019b). Hence, we think the association results presented in this study may be helpful for further investigations.
Table 6Genetic effects of DGAT1 K232A polymorphism on milk yield and milk component traits in Holstein Friesian breed.
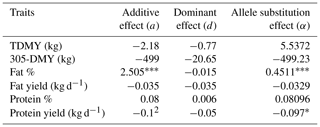
TDMY: test-day milk yield; 305-DMY: 305 d milk yield. Fat and protein yields (kg) were calculated based on TDMY. * p<0.05. p<0.01. p<0.001.
The effect of genotype on FC in the present study was highly significant (p<0.001). The KK genotype had a higher FC (+0.77 % and +0.40 %) than AA and KA. The K allele was associated with higher FC. A recent meta analysis by Mahmoudi and Rashidi (2023) reached similar results for the impact of the K232A marker on FC. These researchers concluded that the K allele tremendously increased milk FC, especially when two copies of this allele are inherited together. In contrast, the A allele of K232A polymorphism had adverse effects. This suggestion which ensued from an extensive statistical aspect by Mahmoudi and Rashidi (2023) has been experimentally substantiated by the present study. However, they found that the K232A marker influenced the PC as well. On the contrary, we observed no statistical difference among the genotypes for the PC in the Holstein Friesian breed (Table 4).
In short, Holstein cows with the AA genotype had higher TDMY (p<0.1) and 305-DMY (p<0.1) but significantly lower FC (p<0.001). Similar results for 305-DMY yields in Holstein cows have also been reported by Sun et al. (2009). Conversely, the same research noted that FC and PC were not different according to genotypes for the DGAT1 marker. The current results agreed with that of Kadlecová et al. (2014) in which AA-genotype animals had the highest milk yield and the lowest FC. Several researchers also pointed out the association between DGAT1 K232A and FC (Carvajal et al., 2016; Li et al., 2021; Manga et al., 2011; Mao et al., 2012; Molee et al., 2015; Schennink et al., 2007; Vanbergue et al., 2016). Unlike the current study, Anton et al. (2012) observed that AA-genotype cows had lower 305-DMY (8247 kg) but high FC (4.34 %) and PC (3.41 %) in Holstein cows. The finding disagrees with the report of Dokso et al. (2015), who observed that the KK genotype (9224.6 kg) was found to be higher than the KA (8866.2 kg) and AA (8825.8 kg) genotypes. However, this difference was not statistically significant. Sun et al. (2009) reported that FC and PC have not differed between genotypes determined for the DGAT1 gene. Tumino et al. (2021) also indicated that K232A polymorphism did not influence the FC. The DGAT1 K232A marker has been shown to be a strong candidate for determining milk production and components. However, the details about the association analysis results and the types of influenced traits may vary widely.
In addition to milk production characteristics in dairy cattle, another critical issue is reproductive performance. Purely yield-oriented selection over many years has resulted in a significant decrease in fertility in dairy cattle. Selection programs supported by current studies include important reproductive characteristics in the indexes (Ardicli et al., 2024; Ooi et al., 2024). Hence, the impact of the DGAT1 gene on bovine reproduction is a crucial assessment in dairy cattle. Rychtarova et al. (2014) and Ardicli et al. (2019b) reported that DGAT1 had a significant effect on calving interval. Demeter et al. (2009) found a potential impact of the DGAT1 on non-return rates for insemination 28 and 56 d after the first service. Collis et al. (2012) found a significant association between the DGAT1 K232A polymorphism and age at puberty. In this study, we evaluated NI and CE in Holstein Friesian cows but did not observe any significant phenotype–genotype relation (Table S1). Berry et al. (2010) also indicated no association of the K232A marker with fertility traits and calving performance in Irish Holstein Friesians. It is important to note that the current knowledge on the influence of DGAT1 K232A marker on reproduction performance is relatively low, especially compared to milk production traits. Hence, it strongly needs further research.
We have observed a salient effect (p<0.001) of DGAT1 K232A polymorphism on the FC in both dairy breeds. Concerning the present analysis, the AA genotype constituted only ∼ 1 % of the total genotype. In many genetic studies, excluding the rare variant is a common implementation in statistical analyses. Hence, we excluded the AA genotype and tested the observed associations subsequently in Holstein Friesians (data not shown). This implementation did not impair the significance of the FC. The dominance effect of DGAT1 K232A for TDMY, FC, and PC was significant in Jersey cows (Table 5). However, there were no significant results for allele substitution effect. A very similar result was presented by Anton et al. (2012), indicating a significant dominance effect on FC and PC in Jersey cows raised in Hungary (p<0.05).
Very recent studies particularly emphasize the importance of this gene in dairy cattle. Overexpression of DGAT1 in adipocytes treated with epinephrine decreased both lipolysis and autophagy, while silencing DGAT1 intensified these processes. Collectively, these results suggest that increasing bovine DGAT1 levels might serve as a regulatory mechanism to inhibit fat breakdown in adipocytes, emphasizing its role in sustaining metabolic balance in dairy cows during energy deficits (Xu et al., 2024). Interestingly, this gene was identified as a positional candidate gene for milk urea (Atashi et al., 2024). The genomic area containing DGAT1 K232A polymorphism is highly dynamic and crucial for several key metabolic processes, including fat and energy balance. In a recent study by Tang et al. (2024), linkage disequilibrium analysis revealed that SNP rs109421300 is closely linked with the K232A mutation in the DGAT1 gene, and it remains an independent gene expression quantitative trait loci (eQTL) even when controlled for in-gene expression studies. Thus, rs109421300 could be a critical marker influencing DGAT1 gene expression, alternative splicing, and various phenotypic traits, warranting further investigation. Archetypal clustering analysis identified a cluster with an archetypal variant near the DGAT1 gene, including variants linked to CDH2, BTRC, SFRP2, ZFHX3, and SLITRK5. This cluster seems to influence milk yield but has minimal impact on fertility. These genes are associated with insulin, adipose tissue, and energy metabolism (Ooi et al., 2024). These studies unequivocally underscore the necessity for further elucidation of critical biological aspects associated with the DGAT1 gene, highlighting the significance of this genomic region in the genetics of dairy cattle.
Regarding the characterization of the genetic effects of the DGAT1 K232A marker on milk yield and milk component traits in the Holstein Friesian breed, the additive effect for FC was highly significant, while no significant dominance effect was observed. The allele substitution effect for FC was also significant (Table 6). This finding was consistent with the results of Koopaei et al. (2012), who observed that the additive effect on FC was significant (+0.455 %; p<0.001). Similarly, Sun et al. (2009) reported the additive effect and allele substitution effect on PY to be 4.67 (p<0.01) and 5.14 (p<0.01). However, they reported no significant additive, dominance, or allele substitution effects for FC and PC. Anton et al. (2012) observed the additive effects on 305-DMY (642.8 kg; p<0.05) and PC (0,091 %; p<0.05). They also reported a significant additive effect (p<0.05) on FC (0.31 %) and a dominance over 305-DMY (189.2 kg), FC (−0.253 %), and PC (−0.045 %). Krovvidi et al. (2021) demonstrated that the least-squares mean of milk yields over lactations was significantly higher (p<0.05) in homozygous (AA) genotypes of both Jersey and Holstein Friesian crossbred cattle after adjusting for farm, parity, and season effects. However, the fat, solids-not-fat (SNF), and protein content in AA genotypes was lower than those in KK genotypes across both genetic groups, though not significantly (p>0.05). Samuel et al. (2023) reported that the AA genotype was linked to higher milk yields in Boran × Holstein Friesian crosses, while the KA genotype was associated with increased yields in zebu population (p<0.05). Milk from KA genotype cattle exhibited lower fat and lactose contents compared to the KK genotype across all genetic groups (p<0.05). Furthermore, replacing one K allele with an A allele significantly reduced daily milk yield by up to 3 L and lactose content by 0.58 %, but increased fat content by up to 0.81 % in the populations studied, underscoring the impactful role of the DGAT1 K232A marker on milk production traits (Samuel et al., 2023).
Our results indicated that the most striking feature of the DGAT1 K232A polymorphism is the highly significant additive effect on the FC. Although previously published reports have pointed out a potential effect of this marker on yield traits, here we report that DGAT1 K232A should be focused on considering milk quality parameters primarily for the fat content of cattle milk. However, it needs further investigation, mainly supported by genotypic interactions and epigenetic dynamics.
This paper extensively focuses on the effects of the DGAT1 K232A polymorphism concerning milk production traits in dairy cattle. Significant differences exist between the genotypes of KA and KK belonging to the DGAT1 marker on test-day milk yield, fat, and protein contents in the Jersey cows. In this breed, the heterozygous genotype had higher test-day milk yield but lower fat and protein contents. Regarding the Holstein Friesian breed, we found significant differences among genotypes on milk fat content. Remarkably, Holstein Friesians cows that possess the KK genotype displayed significantly higher milk fat content means compared to both AA-genotype cows and heterozygotes. Similarly, Jersey cows with the KK genotype demonstrated enhanced milk fat content and, moreover, milk protein content relative to those with the heterozygous genotype. Consequently, we comprehensively investigate the effects of the DGAT1 K232A marker on milk yield and quality in dairy herds of Holstein Friesian and Jersey cows. Consideration of this marker on the development process of the milk quality of cows will provide much more reliable results in a shorter period. This study confirmed the results of some studies in the literature, and particular contradictory results were observed. Furthermore, this study has yielded valuable data for genetic evaluations aimed at enhancing milk content and offers reliable, detailed, and comparative insights into DGAT1 K232A, a critical genetic marker in dairy cattle, across two prevalent dairy breeds.
The datasets generated are available from the corresponding author on request.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-67-455-2024-supplement.
SA: genetics methodology, conceptualization, laboratory experiments, statistical analysis, manuscript writing, and review and editing. OC: data collection, animal management, laboratory experiments, methodology, conceptualization, supervision, and editing. EK: data collection, animal management, laboratory experiments, and methodology. SHA: data collection, animal management, laboratory experiments, and methodology. EKG: data collection, animal management, laboratory experiments, and methodology. SC: data collection, manuscript writing, and methodology.
The contact author has declared that none of the authors has any competing interests.
This research adhered to all pertinent national regulations and institutional protocols concerning the welfare and utilization of animals. Approval was obtained from the Bursa Uludag University Local Ethics Committee for Animal Research (approval number 2010/6-05).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This research has been supported by the Türkiye Bilimsel ve Teknolojik Araştırma Kurumu (grant no. 110O821).
This paper was edited by Henry Reyer and reviewed by Behailu Samuel and two anonymous referees.
Akyuz, B., Agaoglu, O. K., Akçay, A., and Agaoglu, A. R.: Effects of DGAT1 and GH1 polymorphism on milk yield in Holstein cows reared in Turkey, Slov. Vet. Res., 52, 185–191, 2015.
Anton, I., Kovács, K., Fésüs, L., Várhegyi, J., Lehel, L., Hajda, Z., Polgar, J., Szabó, F., and Zsolnai, A.: Effect of DGAT1 and TG gene polymorphisms on intramuscular fat and on milk production traits in different cattle breeds in Hungary, Acta Vet. Hung., 56, 181–186, https://doi.org/10.1556/avet.56.2008.2.5, 2008.
Anton, I., Kovács, K., Holló, G., Farkas, V., Szabó, F., Egerszegi, I., Rátky, J., Zsolnai, A., and Brüssow, K. P.: Effect of DGAT1, leptin and TG gene polymorphisms on some milk production traits in different dairy cattle breeds in Hungary, Arch. Anim. Breed., 55, 307–314, https://doi.org/10.5194/aab-55-307-2012, 2012.
Ardicli, S., Soyudal, B., Samli, H., Dincel, D., and Balci, F.: Effect of STAT1, OLR1, CSN1S1, CSN1S2, and DGAT1 genes on milk yield and composition traits of Holstein breed, Rev. Bras. Zootec., 47, e20170247, https://doi.org/10.1590/rbz4720170247, 2018.
Ardicli, S., Samli, H., Vatansever, B., Soyudal, B., Dincel, D., and Balci, F.: Comprehensive assessment of candidate genes associated with fattening performance in Holstein–Friesian bulls, Arch. Anim. Breed., 62, 9–32, https://doi.org/10.5194/aab-62-9-2019, 2019a.
Ardicli, S., Samli, H., Soyudal, B., Dincel, D., and Balci, F.: Evaluation of candidate gene effects and environmental factors on reproductive performance of Holstein cows, S. Afr. J. Anim. Sci., 49, 379–374, https://doi.org/10.4314/sajas.v49i2.17, 2019b.
Ardicli, S., Aldevir, Ö., Aksu, E., Kucuk, K., and Gümen, A.: Associations of bovine beta-casein and kappa-casein genotypes with genomic merit in Holstein Friesian cattle, Arch. Anim. Breed., 67, 61–71, https://doi.org/10.5194/aab-67-61-2024, 2024.
Ashwell, M., Heyen, D., Sonstegard, T. S., Van Tassell, C. P., Da, Y., VanRaden, P. M., Ron, M., Weller, J. I., and Lewin, H. A.: Detection of quantitative trait loci affecting milk production, health, and reproductive traits in Holstein cattle, J. Dairy Sci., 87, 468–475, https://doi.org/10.3168/jds.S0022-0302(04)73186-0, 2004.
Atashi, H., Chen, Y., Vanderick, S., Hubin, X., and Gengler, N.: Single-step genome-wide association analyses for milk urea concentration in Walloon Holstein cows, J. Dairy Sci., 107, 3020–3031, https://doi.org/10.3168/jds.2023-23902, 2024.
Berry, D. P., Howard, D., O'Boyle, P., Waters, S., Kearney, J. F., and McCabe, M.: Associations between the K232A polymorphism in the diacylglycerol-O-transferase 1 (DGAT1) gene and performance in Irish Holstein-Friesian dairy cattle, Ir. J. Agric. Food Res., 49, 1–9, 2010.
Bobbo, T., Tiezzi, F., Penasa, M., De Marchi, M., and Cassandro, M.: Association analysis of diacylglycerol acyltransferase (DGAT1) mutation on chromosome 14 for milk yield and composition traits, somatic cell score, and coagulation properties in Holstein bulls, J. Dairy Sci., 101, 8087–8091, https://doi.org/10.3168/jds.2018-14533, 2018.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, 1980.
Bovenhuis, H., Visker, M., Poulsen, N., Sehested, J., van Valenberg, H. J. F., van Arendonk, J. A. M., Larsen, L. B., and Buitenhuis, A. J.: Effects of the diacylglycerol o-acyltransferase 1 (DGAT1) K232A polymorphism on fatty acid, protein, and mineral composition of dairy cattle milk, J. Dairy Sci., 99, 3113–3123, https://doi.org/10.3168/jds.2015-10462, 2016.
Carvajal, A., Huircan, P., Dezamour, J., Subiabre, I., Kerr, B., Morales, R., and Ungerfeld, E. M.: Milk fatty acid profile is modulated by DGAT1 and SCD1 genotypes in dairy cattle on pasture and strategic supplementation, Genet. Mol. Res., 15, 15027057, https://doi.org/10.4238/gmr.15027057, 2016.
Collis, E., Fortes, M., Zhang, Y., Tier, B., Schutt, K., Barendse, W., and Hawken, R.: Genetic variants affecting meat and milk production traits appear to have effects on reproduction traits in cattle, Anim. Genet., 43, 442–446, https://doi.org/10.1111/j.1365-2052.2011.02272.x, 2012.
Curi, R. A., Palmieri, D. A., Suguisawa, L., Oliveira, H. N., Silveira, A. C., and Lopes, C. R.: Growth and carcass traits associated with GH1/Alu I and POU1F1/Hinf I gene polymorphisms in Zebu and crossbred beef cattle, Genet. Mol. Biol., 29, 56–61, https://doi.org/10.1590/S1415-47572006000100012, 2006.
Demeter, R. M., Schopen, G. C. B., Oude Lansink, A. G. J. M., Meuwissen, M. P. M., and van Arendonk, J. A. M.: Effects of milk fat composition, DGAT1, and SCD1 on fertility traits in Dutch Holstein cattle, J. Dairy Sci., 92, 11, 5720–5729, https://doi.org/10.3168/jds.2009-2069, 2009.
Dokso, A., Ivanković, A., Zečević, E., and Brka, M.: Effect of DGAT1 gene variants on milk quantity and quality in Holstein, Simmental and Brown Swiss cattle breeds in Croatia, Mljekarstvo, 65, 238–242, https://doi.org/10.15567/mljekarstvo.2015.0403, 2015.
Ensembl genome browser: DGAT1, Cattle Gene, https://www.ensembl.org/index.html, last access: 26 May 2024.
Falconer, D. S. and Mackay, T. F. C.: Introduction to Quantitative Genetics, Harlow, Pearson Education Ltd., ISBN-10: 0582243025, 1996.
Fink, T., Lopdell, T. J., Tiplady, K., Handley, R., Johnson, T. J. J., Spelman, R. J., Davis, S. R., Snell, R. G., and Littlejohn, M. D.: A new mechanism for a familiar mutation–bovine DGAT1 K232A modulates gene expression through multi-junction exon splice enhancement, BMC Genomics, 21, 591, https://doi.org/10.1186/s12864-020-07004-z, 2020.
González-Recio, O., De Maturana, E. L., and Gutiérrez, J.: Inbreeding depression on female fertility and calving ease in Spanish dairy cattle, J. Dairy Sci., 90, 5744–5752, https://doi.org/10.3168/jds.2007-0203, 2007.
Gothwal, A., Magotra, A., Bangar, Y. C., Malik, B. S., Yadav, A. S., and Garg, A. R.: Candidate K232A mutation of DGAT1 gene associated with production and reproduction traits in Indian Dairy cattle, Anim. Biotechnol., 34, 2608–2616, https://doi.org/10.1080/10495398.2022.2109041, 2023.
Green, M. J. S.: Isolation of high-molecular-weight DNA from mammalian cells using proteinase K and phenol, in: Molecular Cloning: A Laboratory Manual, 4th Edn., Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press, 47–48, ISBN-13: 978-1936113422, 2012.
Grisart, B, Coppieters, W, Farnir, F, Karim, L., Ford, C., Berzi, P., Cambisano, N., Mni, M., Reid, S., Simon, P., Spelman, R., Georges, M., and Snell, R.: Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition, Genome Res., 12, 222–231, https://doi.org/10.1101/gr.224202, 2002.
Grisart, B., Farnir, F., Karim, L., Cambisano, N., Kim, J. J., Kvasz, A., Mni, M., Simon, P., Frere, J.-M., Coppieters W., and Georges, M.: Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition, P. Natl. Acad. Sci. USA, 101, 2398–2403, 2004.
Kadlecová, V., Nĕmĕcková, D., Ječmínková, K., and Stádník, L.: Association of bovine DGAT1 and leptin genes polymorphism with milk production traits and energy balance indicators in primiparous Holstein cows, Mljekarstvo/Dairy, 64, 19–26, ISSN 1846-4025, Hrcak ID 117066, 2014.
Kaupe, B., Winter, A., Fries, R., and Erhardt, G.: DGAT1 polymorphism in Bos indicus and Bos taurus cattle breeds, J. Dairy Res., 71, 182–187, 2004.
Khan, M. Z., Wang, D., Liu, L., Usman, T., Wen, H., Zhang, R., Liu, S., Shi, L., Mi, S., Xiao, W., and Yu, Y.: Significant genetic effects of JAK2 and DGAT1 mutations on milk fat content and mastitis resistance in Holsteins, J. Dairy Res., 86, 388–393, https://doi.org/10.1017/S0022029919000682, 2019.
Komisarek, J., Waskowicz, K., Michalak, A., and Dorynek, Z.: Effects of DGAT1 variants on milk production traits in Jersey cattle, Anim. Sci. Pap. Rep., 22, 307–313, 2004.
Kong H, Oh J, Lee J, Yoon, D. H., Choi, Y. H., Cho, B. W., Lee, H. K., and Jeon, G. J.: Association of sequence variations in DGAT 1 gene with economic traits in Hanwoo (Korea cattle), Asian-Australas, J. Anim. Sci., 20, 817–820, https://doi.org/10.5713/ajas.2007.817, 2007.
Koopaei, H. K., Abadi, M. R. M., Mahyari, S. A., Tarang, A. R., Potki, P., and Koshkoiyeh, A. E.: Effect of DGAT1 variants on milk composition traits in Iranian Holstein cattle population, Anim. Sci. Pap. Rep., 30, 231–239, 2012.
Krovvidi, S., Thiruvenkadan, A. K., Murali N., Saravanan, R., Vinoo, R., and Metta, M.: Evaluation of non-synonym mutation in DGAT1 K232A as a marker for milk production traits in Ongole cattle and Murrah buffalo from Southern India, Trop. Anim. Health Prod., 53, 118, https://doi.org/10.1007/s11250-021-02560-2, 2021.
Lehner, R. and Kuksis A.: Biosynthesis of triacylglycerols, Prog. Lipid Res., 35, 169–201, https://doi.org/10.1016/0163-7827(96)00005-7, 1996.
Lešková, L., Bauer, M., Chrenek, P., Lackova, Z., Soročinová, J., Petrovič, V., and Kováč, G.: Detection of DGAT1 gene polymorphism and its effect on selected biochemical indicators in dairy cows after calving, Acta Vet. Brno, 82, 265–269, https://doi.org/10.2754/avb201382030265, 2013.
Li, Y., Zhou, H., Cheng, L., Edwards, G. R., and Hickford, J. G. H.: Effect of DGAT1 variant (K232A) on milk traits and milk fat composition in outdoor pasture-grazed dairy cattle, N. Z. J. Agric. Res., 64, 101–113, https://doi.org/10.1080/00288233.2019.1589537, 2021.
Loftus, R. T., MacHugh, D. E., Bradley, D. G., Sharp, P. M., and Cunningham, P.: Evidence for two independent domestications of cattle, P. Natl. Acad. Sci. USA, 91, 2757–2761, 1994.
Magan, J. B., O'Callaghan, T. F., Kelly, A. L., and McCarthy, N. A.: Compositional and functional properties of milk and dairy products derived from cows fed pasture or concentrate-based diets, Compr. Rev. Food Sci. F., 20, 2769–2800, https://doi.org/10.1111/1541-4337.12751, 2021.
Mahmoudi, P. and Rashidi, A.: Strong evidence for association between K232A polymorphism of the DGAT1 gene and milk fat and protein contents: A meta-analysis, J. Dairy Sci., 106, 2573–2587, https://doi.org/10.3168/jds.2022-22315, 2023.
Manga, I. and Říha, H.: The DGAT1 gene K232A mutation is associated with milk fat content, milk yield and milk somatic cell count in cattle, Arch. Anim. Breed., 54, 257–263, https://doi.org/10.5194/aab-54-257-2011, 2011.
Mansbridge, R. and Blake, J.: Nutritional factors affecting the fatty acid composition of bovine milk, Br. J. Nutr., 78, S37–S47, https://doi.org/10.1079/BJN19970133, 1997.
Mao Y, Chen R, Chang L, Chen, Y., Ji, D. J., Wu, X. X., Shi, X. K., Wu., H. T., Zhang, M. R., Yang, Z. P., König, S., and Yang, L. G.: Effects of SCD1-and DGAT1-genes on production traits of Chinese Holstein cows located in the Delta Region of Yangtze River, Livest. Sci., 145, 280–286, https://doi.org/10.1016/j.livsci.2011.12.019, 2012.
Medjugorac I, Kustermann W, Lazar P, Russ, I., and Pirchner, F.: Marker-derived phylogeny of European cattle supports demic expansion of agriculture, Anim. Genet., 25, 19–27, https://doi.org/10.1111/j.1365-2052.1994.tb00399.x, 1994.
Mohammed, S.A., Rahamtalla, S. A., Ahmed, S. S., Elhafiz, A., Dousa, B. M., Elamin, K. M., and Ahmed, M. K. A.: DGAT1 gene in dairy cattle, Glob. J. Anim., 3, 191–198, 2015.
Molee, A., Poompramun, C., and Mernkrathoke, P.: Effect of casein genes-beta-LGB, DGAT1, GH, and LHR-on milk production and milk composition traits in crossbred Holsteins, Genet. Mol. Res., 14, 2561–2571, https://doi.org/10.4238/2015.March.30.15, 2015.
Nei, M. and Roychoudhury, A: Sampling variances of heterozygosity and genetic distance, Genetics, 76, 379–390, 1974.
Ooi, E., Xiang, R., Chamberlain, A. J., and Goddard, M. E.: Archetypal clustering reveals physiological mechanisms linking milk yield and fertility in dairy cattle, J. Dairy Sci., 107, 4726–4742, https://doi.org/10.3168/jds.2023-23699, 2024.
Pathak, R. K., Lim, B., Park, Y., and Kim, J. M.: Unraveling structural and conformational dynamics of DGAT1 missense nsSNPs in dairy cattle, Sci. Rep., 12, 4873, https://doi.org/10.1038/s41598-022-08833-6, 2022.
Ripoli, M. V., Corva, P., and Giovambattista, G.: Analysis of a polymorphism in the DGAT1 gene in 14 cattle breeds through PCR-SSCP methods, Res. J. Vet. Sci., 80, 287–290, https://doi.org/10.1016/j.rvsc.2005.07.006, 2006.
Rychtarova, J., Sztankoova, Z., Kyselova, J., Zink, V., Stipkova, M., Vacek, M., and Stolc, L.: Effect of DGAT1, BTN1A1, OLR1, and STAT1 genes on milk production and reproduction traits in the Czech Fleckvieh breed, Czech J. Anim. Sci., 59, 45–53, 2014.
Samuel, B., Dadi, H., and Dinka, H.: Effect of the DGAT1 K232A mutation and breed on milk traits in cattle populations of Ethiopia, Front. Anim. Sci., 4, 1096706, https://doi.org/10.3389/fanim.2023.1096706, 2023.
Schennink, A., Stoop, W. M., Visker, M. W., Heck, J. M. L., Bovenhuis, H., Van Der Poel, J. J., Van Valenberg, H. J. F., and Van Arendonk, J. A. M.: DGAT1 underlies large genetic variation in milk-fat composition of dairy cows, Anim. Genet., 38, 467–473, https://doi.org/10.1111/j.1365-2052.2007.01635.x, 2007.
Strzałkowska, N., Siadkowska, E., Słoniewski, K., Krzyżewski, J., and Zwierzchowski, L.: Effect of the DGAT1 gene polymorphism on milk production traits in Black-and-White (Friesian) cows, Anim. Sci. Pap. Rep., 23, 189–197, 2005.
Sun, D., Jia, J., Ma, Y., Zhang, Y., Wang, Y., Yu, Y., and Zhang, Y.: Effects of DGAT1 and GHR on milk yield and milk composition in the Chinese dairy population, Anim. Genet., 40, 997–1000, https://doi.org/10.1111/j.1365-2052.2009.01945.x, 2009.
Tang, Y., Zhang, J., Li, W., Liu, X., Chen, S., Mi, S., Yang, J., Teng, J., Fang, L., and Yu, Y.: Identification and characterization of whole blood gene expression and splicing quantitative trait loci during early to mid-lactation of dairy cattle, BMC Genomics, 25, 445, https://doi.org/10.1186/s12864-024-10346-7, 2024.
Thaller, G., Kramer, W., Winter, A., Kaupe, B., Erhardt, G., and Fries, R.: Effects of DGAT1 variants on milk production traits in German cattle breeds, J. Anim. Sci., 81, 1911–1918, https://doi.org/10.2527/2003.8181911x, 2003.
Tumino S, Criscione A, Moltisanti V, Marletta, D., Bordonaro, S., Avondo, M., and Valenti, B.: Feeding system resizes the effects of dgat1 polymorphism on milk traits and fatty acids composition in modicana cows, Animals, 11, 1616, https://doi.org/10.3390/ani11061616, 2021.
Valenti, B., Criscione, A., Moltisanti, V., Bordonaro, S., De Angelis, A., Marletta, D., Di Paola, F., and Avondo, M.: Genetic polymorphisms at candidate genes affecting fat content and fatty acid composition in Modicana cows: effects on milk production traits in different feeding systems, Animal, 13, 1–9, https://doi.org/10.1017/S1751731118002604, 2019.
Vanbergue, E., Peyraud, J.-L., Guinard-Flament, J., Charton, C., Barbey, S., Lefebvre, R., Gallard, Y., and Hurtaud, C.: Effects of DGAT1 K232A polymorphism and milking frequency on milk composition and spontaneous lipolysis in dairy cows, J. Dairy Sci., 99, 5739–5749, https://doi.org/10.3168/jds.2015-10731, 2016.
Van Gastelen, S., Visker, M., Edwards, J., Antunes-Fernandes, E. C., Hettinga, K. A., Alferink, S. J. J., Hendriks, W. H., Bovenhuis, H., Smidt, H., and Dijkstra, J.: Linseed oil and DGAT1 K232A polymorphism: Effects on methane emission, energy and nitrogen metabolism, lactation performance, ruminal fermentation, and rumen microbial composition of Holstein-Friesian cows, J. Dairy Sci., 100, 8939–8957, https://doi.org/10.3168/jds.2016-12367, 2017.
Xu, Q., Fan, Y., Mauck, J., Loor, J. J., Sun, X., Jia, H., Li, X., and Xu, C.: Role of diacylglycerol O-acyltransferase 1 (DGAT1) in lipolysis and autophagy of adipose tissue from ketotic dairy cows, J. Dairy Sci., 107, 5150–5161, https://doi.org/10.3168/jds.2023-24471, 2024.
Weller, J., Golik, M., Seroussi, E., Ezra, E., and Ron, M.: Population-wide analysis of a QTL affecting milk-fat production in the Israeli Holstein population, J. Dairy Sci., 86, 2219–2227, https://doi.org/10.3168/jds.S0022-0302(03)73812-0, 2003.
Winter, A., Krämer, W., Werner, F. A., Kollers, S., Kata, S., Durstewitz, G., Buitkamp, J., Womack, J. E., Thaller, G., and Fries, R.: Association of a lysine-232 alanine polymorphism in a bovine gene encoding acyl-CoA: diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content, P. Natl. Acad. Sci. USA, 99, 9300–9305, 2002.





