the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The role of miR-369-3p in proliferation and differentiation of preadipocytes in Aohan fine-wool sheep
Shijie Xue
Kaidong Liu
Le Zhao
Lisheng Zhou
Xiaoxiao Gao
Lirong Liu
Nan Liu
Jianning He
MicroRNAs (miRNAs) are a large class of non-coding RNAs that play important roles in the proliferation and differentiation of adipocytes. Our previous sequencing analysis revealed higher expression of miR-369-3p in the longissimus muscle of 2-month-old Aohan fine-wool sheep (AFWS) compared to 12-month-old sheep (P<0.05), suggesting that miR-369-3p may regulate fat deposition in AFWS. To test this, miR-369-3p mimics, inhibitors, and negative controls (NCs) were constructed and transfected into AFWS preadipocytes. After transfection with miR-369-3p mimics, we found a decrease (P<0.05) in the expression of genes and proteins related to cell proliferation and differentiation, detected by RT-qPCR (quantitative reverse transcription PCR) and western blot analyses. Moreover, EdU (5-ethynyl-2′-deoxyuridine) detection and Oil Red O staining showed a decrease (P<0.05) in cell proliferation and lipid accumulation, respectively. The opposite trends (P<0.05) were obtained after transfection with miR-369-3p inhibitors. In conclusion, the results showed that miR-369-3p can inhibit the proliferation and differentiation of AFWS preadipocytes, providing a theoretical basis to further explore the molecular mechanism of fat deposition in sheep and other domestic animals.
- Article
(2488 KB) - Full-text XML
-
Supplement
(518 KB) - BibTeX
- EndNote
Mutton is becoming increasingly popular as living standards improve and dietary patterns change because of its unique taste, high protein content, and low cholesterol content (Jia et al., 2023). Intramuscular fat (IMF) content is a key indicator of meat quality, affecting meat colour, flavour, tenderness, and other traits, and increasing IMF relative to other indicators can improve the quality of meat (Mortimer et al., 2017). Therefore, determining how to improve IMF content is essential to improving the quality of meat.
MicroRNAs (miRNAs) are a class of 20–25 nt non-coding RNAs that regulate gene expression by targeting mRNA (Iorio et al., 2005; Yekta et al., 2004). Many miRNAs have been reported as regulators of fat formation and metabolism. For example, miR-145 downregulates ADAM17 to promote the decomposition of human adipocytes (Lorente-Cebrián et al., 2014), miR-182 and miR-203 act in the differentiation of brown adipocytes (Kim et al., 2014), and miR-155 directly targets and inhibits PPARγ expression to regulate adipocytes (Karkeni et al., 2016). Studies relating to sheep show that miR-27a regulated the formation of lipid droplets in sheep adipocytes by targeting the CPT1B gene (Li et al., 2021), and miR-340-5p inhibits sheep adipocyte differentiation by targeting ATF7 (Liu et al., 2020). In a preliminary study, we evaluated the IMF content of 2-, 4-, 6-, and 12-month-old Aohan fine-wool sheep (AFWS). The results showed that the IMF content in the 2- and 12-month-old sheep was significantly different (Han et al., 2021a). Then we found many differentially expressed miRNAs in back longissimus muscle between 2-month- and 12-month-old AFWS by RNA-seq (Han et al., 2021b). Of these miRNAs, the expression level of miR-369-3p at 2 months old was significantly higher than at 12 months old (Table S1 in the Supplement). Therefore, miR-369-3p may have a regulatory effect on intramuscular fat content.
There have been few studies regarding the regulation of miR-369-3p on fat, whereas the majority of studies focused on disease. Galleggiante et al. (2019) showed that miR-369-3p inhibited the expression of C/EBPβ. C/EBPβ is the first set of transcription factors regulating the process of fat formation. Mouse embryonic fibroblasts (3T3-L1) lacking these genes cannot complete the differentiation of fat induced by hormones and interfere with normal expression of other adipogenic genes. C/EBPβ also promotes cell mitosis in the process of fat formation. When induced in vitro, precursor adipocytes reenter the cell division cycle at the early stage of differentiation, and C/EBPβ promotes cell mitosis in this process, which is required for adipocyte differentiation (Azad et al., 2014). The binding of miR-369-3p to the 3′UTR region of TSPAN13 mRNA inhibited expression of TSPAN13, leading to a decrease in CDK4 expression (Li et al., 2019; Lou et al., 2017). CDK4 indirectly inhibits oxidative metabolism by controlling E2F1 transcription factors in muscle and brown adipose tissue (BAT) (Blanchet et al., 2011) and also promotes insulin signalling pathways in mature adipocytes (Lagarrigue et al., 2016). In sheep, miR-369-3p targets the key gene PDPK-1 that activates autophagy, and its expression is negatively correlated (Lie et al., 2016; Hu et al., 2021). The miR-369-3p can also regulate and inhibit the autophagy-related gene ATG10 (Liu et al., 2019).
Therefore, a significant amount of data has shown that miR-369-3p is closely related to genes related to adipocyte proliferation and differentiation. To further investigate the role of miR-369-3p in AFWS, AFWS preadipocytes were used to study the effect of miR-369-3p on preadipocyte proliferation and differentiation.
2.1 Materials
Samples of preadipocytes, back longissimus muscle, biceps femoris, shoulder skin, pituitary, heart, lung, testicle, liver, and small intestine were collected from 4 d old Aohan fine-wool male lambs obtained from Aohan Breeding Farm in the city of Chifeng, Inner Mongolia. We anesthetized the sheep by intravenous injection of sodium pentobarbital at a dose of 25 mg kg−1 following published protocols (Raj et al., 2004; Hawkins et al., 2016) and then placed it in a closed room that was filled with carbon dioxide at a rate of 20 % min−1. When the gas concentration had reached 80 %, the sheep died.
2.2 Differentially expressed miRNAs in sheep muscle tissue
In a preliminary study, we found many differentially expressed miRNAs in back longissimus muscle between 2-month- and 12-month-old AFWS by RNA-seq (Han et al., 2021b). Of these miRNAs, the expression level of miR-369-3p at 2 months old was significantly higher than at 12 months old (Table S1).
2.3 Culture and passage of AFWS preadipocytes
After the 4 d old AFWS was killed, the longissimus muscle tissue was taken and washed five times with PBS (phosphate-buffered saline)) containing 0.1 % penicillin streptomycin. We also removed the visible blood vessels and connective tissue in sterile culture dishes. Then we cut the longissimus muscle tissue into a tissue block of about 1.0 mm, adding 0.2 % collagenase II (Solarbio, Beijing, China) before putting the tissue into a 37 ∘C water bath for digestion in 1.5 h. Shaking it every 5 min, we added a complete medium (Dulbecco's Modified Eagle Medium (DMEM) + 10 % fetal bovine serum (FBS) + 0.1 % penicillin streptomycin) of equal volume after digestion. Digestive solution was filtered with 200 mesh and 400 mesh cell sieves, after which the cells were resuspended in the complete medium and placed in the CO2 incubator for 3–4 h. The adherent cells were preadipocytes. When the confluence degree of primary cell culture reached 80 %, the cells were passaged at a ratio of 1 : 2 and placed in a 37 ∘C, 5 % CO2 incubator.
2.4 Induction of preadipocyte differentiation
After 2 d of fusion, the cells were induced in the first stage after high cell fusion (DMEM, FBS, penicillin streptomycin, methylisobutylxanthine, dexamethasone, insulin). Before induction, the cells were collected for RNA and protein extraction, which was recorded as day 0. After the first stage, some cells became round. After 2 d, the second stage of induction (DMEM, FBS, insulin, penicillin streptomycin) was conducted. The culture medium was changed every 2 d, with cell collection.
2.5 Transfection
After the cells were digested using trypsin, they were evenly distributed in a six-well plate with 2 mL of complete medium and cultured overnight. When the density reached about 60 %, the mimics, inhibitors, and negative controls (NCs) (Jima Gene, Shanghai, China) were transfected into the cells with GP-transfect-Mate (Jima Gene, Shanghai, China) according to the manufacturer's instructions (A: miR-369-3p mimic, B: miR-369-3p mimic NC, C: miR-369-3p inhibitor, D: miR-369-3p NC). After 48 h, the cells were collected to detect the expression of miR-369-3p.
2.6 RNA extraction and reverse transcription
RNA was extracted from back longissimus muscle, biceps femoris, shoulder skin, pituitary, heart, lung, testis, spleen, and small intestine of 4 d old AFWS lambs. After grinding, total RNA was extracted using a SPARKeasy total RNA rapid extraction kit (Shandong Sikejie Biotechnology Co., Ltd.). The cDNA was prepared from the RNA using an Evo M-MLV reverse transcription kit (Aikerui Biotechnology, Hunan, China), followed by an miRNA cDNA first island chain synthesis kit (Aikerui Biotechnology, Hunan, China).
2.7 RT-qPCR (quantitative reverse transcription PCR)
The expression levels of mRNAs and miRNA were quantified by the CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States). Primers were designed using Primer 5 (Table S2) and synthesized by Tsingke Biological Technology (Tsingke, Qingdao, China). Reverse transcription was conducted using 1 µg total RNA per sample and the Mir-X miRNA First-Strand Synthesis Kit (TaKaRa, Dalian, China). The levels of U6 and GAPDH were used to normalize the expression levels of mRNA and miRNAs, respectively. The sequences of the primers used are listed in Table 1. The 20 µL PCR reaction system consisted of 10 µL SYBR® Premix Ex Taq II (TaKaRa), 0.5 µL forward primer (10 µM L−1), 0.5 µL reverse primer (10 µM L−1), 1 µL cDNA, and 8 µL ddH2O. The following thermocycling programme was used: one cycle of 95 ∘C for 10 min; 45 cycles of 95 ∘C for 10 s, 60 ∘C for 10 s, and 72 ∘C for 10 s; and one cycle of 72 ∘C for 6 min. Three independent replicates were conducted for each sample. The relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.8 Total protein extraction and western blot analysis
Total proteins were extracted using RIPA Lysis Buffer (Beyotime, Shanghai, China), supplemented with phenylmethylsulfonyl fluoride (Service-Bio, Wuhan, China) at a ratio of 100 : 1 on ice. The extracted protein concentration was then measured by a BCA Protein Assay Kit (Solarbio, Beijing, China) (Gao et al., 2019). Proteins (30 µg) were separated by 10 % SDS-PAGE polyacrylamide gel electrophoresis and then transferred onto PVDF (polyvinylidene difluoride) membranes (Beyotime). The membranes were blocked with 5 % skimmed milk for 2 h and then incubated overnight at 4 ∘C with rabbit anti-PPARγ, rabbit anti-C/EBPα, rabbit anti-CDK4, or rabbit anti-CyclinB at a dilution of 1 : 2000. The membranes were then incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (1 : 2000) for 1 h at room temperature, followed by washing. Rabbit anti-GAPDH was used as an internal control. All antibodies were purchased from Proteintech (Chicago, IL, United States). The protein signals were detected in a darkroom using a Pierce ECL Western Blotting Substrate kit (Thermo Fisher Scientific, Dallas, TX, United States) according to the manufacturer's instructions.
2.9 Oil Red O staining
Differentiation was induced in preadipocytes for 8 d, and then the cells were stained with Oil Red O (ORO; Solarbio, Beijing, China). The culture medium was removed, and cells were washed with PBS (phosphate buffered saline) and fixed with ORO fixative for 30 min. The fixative was removed, cells were rinsed with distilled water twice, and 60 % isopropanol was added and incubated for 5 min before removal. The ORO stain was then added and incubated for 15 min before removal. The stained cells were washed five times with distilled water. After washing, Mayer's hematoxylin staining solution was added, and cells were re-stained for 2 min. After discarding, the stained cells were washed five times with distilled water. ORO buffer was added, incubated for 1 min, and then discarded. Finally, the cells were covered with distilled water and observed under a microscope (Leica Microsystems, Wetzlar, Germany).
2.10 EdU detection of cell proliferation
After passage to the six-well plate, preadipocytes in good condition and at the growth density of 70 % were selected for EdU (5-ethynyl-2′-deoxyuridine) staining using an EdU cell proliferation detection kit (Beyotime, Shanghai, China). Cells were fixed with 4 % paraformaldehyde and labelled with EdU, and then the nuclei were stained using Hoechst 33342.
2.11 Statistical analysis
IBM SPSS Statistics 26.0 software was used to calculate the relative expression level of genes using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The relative expression of genes was analysed by an independent sample t test, and the results were expressed as mean ± SE (standard error). A value of P<0.05 was considered a significant difference.
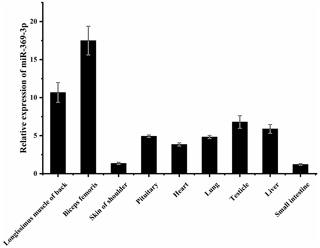
3.1 Expression of miR-369-3p in different AFWS tissues
To explore whether miR-369-3p mainly plays a role in sheep muscle, its expression on different AFWS tissues was determined. As shown in Fig. 1, RT-qPCR showed that miR-369-3p was widely expressed in various tissues, with higher expression levels in the back longissimus muscle and biceps femoris. As shown in Fig. S1 in the Supplement, comparison of the miR-369-3p sequences of 14 species revealed high conservation, indicating these miRNAs may play important roles.
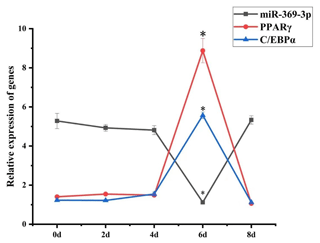
3.2 The expression of miR-369-3p in preadipocyte differentiation
To explore the role of miR-369-3p in AFWS preadipocyte differentiation, its expression on consecutive days following induction of differentiation was determined. The results of RT-qPCR are shown in Fig. 2. The expression levels of marker genes for adipocyte differentiation, PPARγ and C/EBPα, peaked at 6 d, and the differences in expression level were significant (P<0.05). The expression levels decreased after 6 d, indicating a critical period for the differentiation of preadipocytes at 6 d, but the expression level of miR-369-3p reached the lowest value on the sixth day, with a significant difference.
3.3 Transfection effect of the miR-369-3p mimic and inhibitor
To further investigate the expression of miR-369-3p, we transfected cells with a mimic or inhibitor, as well as negative controls (NCs), as shown in Fig. S2. After 48 h of transfection, the expression of miR-369-3p in the mimic group was higher compared to the mimic NC group, and the difference was significant (P<0.05). The expression of miR-369-3p in the inhibitor group was lower compared to the NC group, and this difference was significant (P<0.05). The observed changes in expression indicated successful transfection of the miR-369-3p mimic and inhibitor, allowing for subsequent experiments.
3.4 The expression changes of preadipocyte proliferation and differentiation marker genes after transfection
To further investigate the role of miR-369-3p, the mimic or inhibitor constructs were used, and the effects on the expression levels of preadipocyte proliferation and differentiation marker genes were determined. As shown in Fig. 3, after transfection of the miR-369-3p mimic for 48 h, the expression levels of CDK4, CyclinB, and PPARγ and of C/EBPα were significantly decreased (P<0.05). However, the opposite results were obtained after transfection of the miR-369-3p inhibitor (P<0.05).
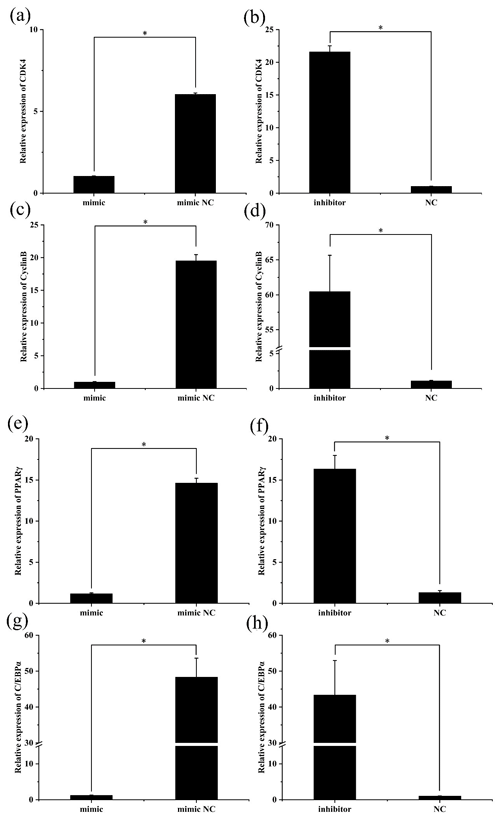
Figure 3The expression levels of preadipocyte proliferation and differentiation marker genes after transfection. (a) Relative expression of CDK4 after overexpression of miR-369-3p. (b) Relative expression of CDK4 after inhibition of miR-369-3p. (c) Relative expression of CyclinB after overexpression of miR-369-3p. (d) Relative expression of CyclinB after inhibition of miR-369-3p. (e) Relative expression of PPARγ after overexpression of miR-369-3p. (f) Relative expression of PPARγ after inhibition of miR-369-3p. (g) Relative expression of C/EBPα after overexpression of miR-369-3p. (h) Relative expression of C/EBPα after inhibition of miR-369-3p (* P<0.05).
3.5 Changes of protein expression after transfection
To further explore the effect of miR-369-3p on the proliferation and differentiation of adipocytes, the total protein was extracted from preadipocytes after transfection for 48 h in each transfected group. The protein expression levels of adipocyte marker genes, CDK4, CyclinB, and PPARγ, as well as C/EBPα, were determined, using the level of GAPDH as a control, as shown in Figs. 4 and 5. The CDK4, CyclinB, and PPARγ as well as C/EBPα protein expression was inhibited by miR-369-3p mimics and promoted by miR-369-3p inhibitors. The results showed changes in adipocyte marker genes consistent with the observed trends in RNA expression detected by RT-qPCR.
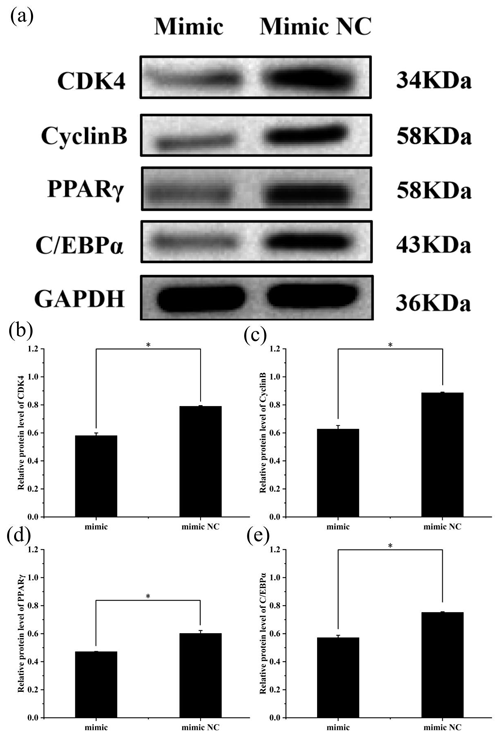
Figure 4Effect of overexpression of miR-369-3p on the expression level of CDK4, CyclinB, PPARγ, and C/EBPα. (a) Western blot analysis of adipogenic genes. (b) Relative protein level of CDK4. (c) Relative protein level of CyclinB. (d) Relative protein level of PPARγ. (e) Relative protein level of C/EBPα (* P<0.05).
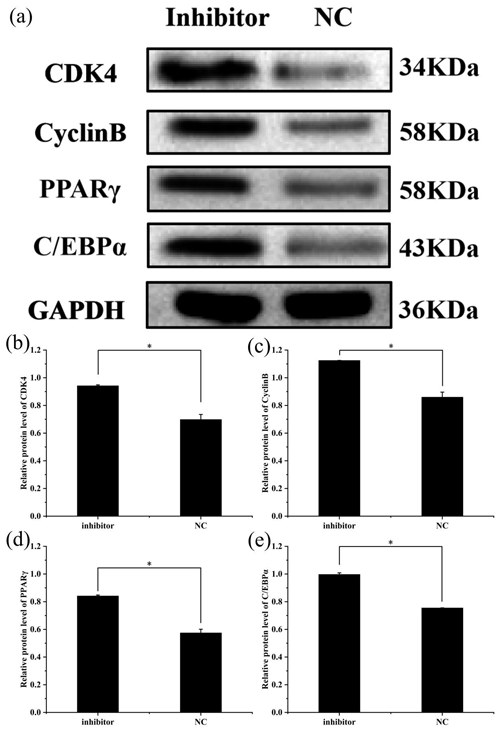
Figure 5Effect of inhibition of miR-369-3p on the expression level of CDK4, CyclinB, PPARγ, and C/EBPα. (a) Western blot analysis of adipogenic genes. (b) Relative protein level of CDK4. (c) Relative protein level of CyclinB. (d) Relative protein level of PPARγ. (e) Relative protein level of C/EBPα (* P<0.05).
3.6 EdU detection of cell proliferation
To assess cell proliferation, EdU detection was performed using the transfected cells, and the results are shown in Fig. 6. Cell proliferation was significantly greater in the miR-369-3p inhibition group compared to the NC group, and the opposite result was obtained in the mimic group (P<0.05). Thus, miR-369-3p acts to inhibit the proliferation of preadipocytes.
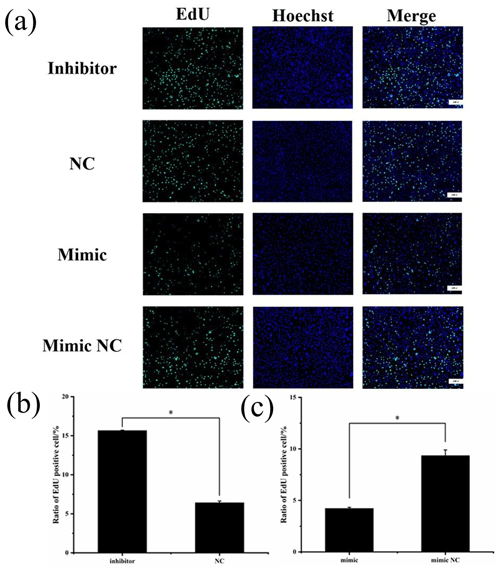
Figure 6(a) EdU detection of cell proliferation after transfection. Blue fluorescence represents the nucleus stained with Hoechst, and green fluorescence represents proliferating cells. (b) Ratio of EdU-positive cell of inhibitor and NCs. (c) Ratio of EdU-positive cell of mimic and mimic NCs (* P<0.05).
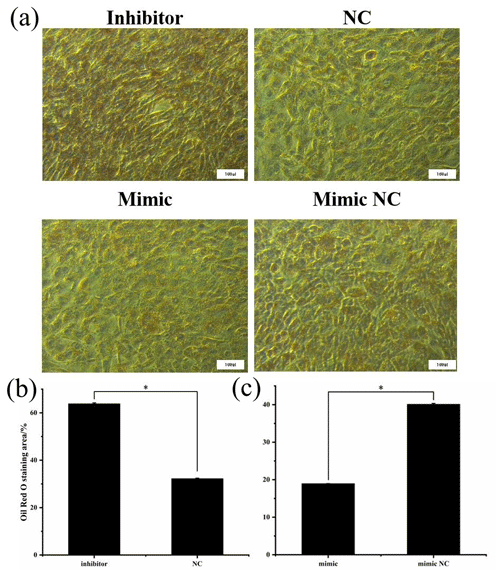
3.7 Oil Red O staining
After being induced for 8 d, the cells were stained with Oil Red O to investigate the effect of miR-369-3p on lipid deposition, as shown in Fig. 7. There was significantly greater staining in the miR-369-3p inhibitor group compared to the NC group, with the opposite results in the mimic group (P<0.05). These results further verified that miR-369-3p can inhibit the differentiation of preadipocytes.
As non-coding RNAs, miRNAs play important roles in a variety of tissues or organs by regulating gene expression. In adipose tissue, miRNAs can regulate adipocyte proliferation, differentiation, metabolism, and endocrine function (Arner and Kulyté, 2015). Our previous work revealed significantly lower expression of miR-369-3p in the longissimus dorsi muscle of 12-month-old AFWS compared to that of 2-month-old sheep (Han et al., 2021b) (Table S1). We found that the expression of miR-369-3p in muscle was higher compared to other tissues. This suggests that miR-369-3p acts in the growth and development of muscles. So far, studies on miR-369-3p have mainly focused on diseases, including cancer and inflammation (Chen et al., 2021; Scalavino et al., 2020; Xu and Liu, 2020). For example, in a study of chronic inflammation, Galleggiante et al. (2019) found that the upregulation of miR-369-3p could inhibit the expression of C/EBPβ, a key transcription factor in the differentiation of preadipocytes (Cao et al., 2017). There was another study showing that during 3T3-L1 adipocyte differentiation, C/EBPβ is induced early to transactivate the expression of C/EBPα and PPARγ (Guo et al., 2015). Members of the C/EBP and PPAR families of transcription factors are the most notable in the differentiation of preadipocytes into adipocytes. Furthermore, C/EBPα and PPARγ are critical regulators of adipogenesis, since the lack of either of these proteins prevents the development of white adipose tissue in mice (Linhart et al., 2001; Rosen et al., 1999; Barak et al., 1999). Therefore, miR-369-3p may inhibit the expression of C/EBPα and PPARγ by inhibiting the expression of C/EBPβ, thus inhibiting the differentiation of adipocytes.
Autophagy removes multiple cellular components during fat differentiation (Mizushima and Levine, 2010). Xu et al. (2018) showed that the expression of the autophagy pathway was negatively correlated with fat decomposition. ATG10 plays an important role in the formation and extension of autophagosomes (Kim and Lee, 2014). Liu et al. (2019) found that miR-369-3p regulates the proliferation and migration of EEC (endometrioid adenocarcinoma) cells, suggesting that miR-369-3p can also inhibit adipocyte differentiation by inhibiting the expression of ATG10 in human body. Lie et al. (2016) showed that miR-369-3p targeted PDPK-1 (3-phosphoinositide-dependent protein kinase-1) in sheep, and expression levels were negatively correlated. PDPK-1 is the key gene to initiate autophagy (Hu et al., 2021), and the expression of PDPK-1 and PPARγ resulted in a synergistic effect (Yin et al., 2006). Hence, miR-369-3p may inhibit the autophagy process and the expression of PPARγ by inhibiting the expression of PDPK-1 in sheep to negatively affect the differentiation of adipose precursor cells.
Lou et al. (2017) found that in CRC (colorectal cancer) cell lines, knocking down TSPAN13 led to the decrease of CDK4 expression, and miR-369-3p inhibited TSPAN13 by binding to the 3'UTR region of TSPAN13 mRNA (Li et al., 2019). Therefore, miR-369-3p likely inhibits the expression of CDK4 by downregulating TSPAN13. In previous studies, overexpression of CDK4 has been shown to promote cell proliferation by driving cell cycle progression (Retzer-Lidl et al., 2007; Rodriguez-Puebla et al., 2002; An et al., 1999). Further, the proper regulation of CyclinB is essential for the entry into mitosis (Krek and Nigg, 1991), and reducing the time between cell birth and mitosis increases the rate of cell multiplication (Moxnes et al., 2004).
In summary, miR-369-3p negatively regulates proliferation and differentiation of AFWS preadipocytes.
In this study, we found that miR-369-3p inhibits the proliferation and differentiation of AFWS preadipocytes. This study provides a theoretical basis for further exploration of the molecular mechanism of fat deposition in sheep and other livestock.
| miRNAs | microRNAs |
| AFWS | Aohan fine-wool sheep |
| NC | negative control |
| IMF | intramuscular fat |
Additional data can be found in the Supplement.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-66-93-2023-supplement.
SX, KL, JH, and NL: conceptualization. SX, KL, LeZ, LiZ, XG, LL, NL, and JH: methodology. SX: software. SX and JH: validation. SX and NL: formal analysis. SX: investigation. SX, KL, LeZ, LiZ, XG, LL, NL, and JH: resources. SX, KL, LeZ, LiZ, XG, LL, NL, and JH: data curation. SX and KL: writing – original draft preparation. SX, KL, and JH: writing – review and editing. SX: visualization. LiZ, JH, and NL: supervision. KL: project administration. NL, LiZ, and JH: funding acquisition. All authors have read and agreed to the published version of the manuscript.
The contact author has declared that none of the authors has any competing interests.
The animal study was reviewed and approved by the Animal Administration and Ethics Committee of Qingdao Agricultural University, Animal Science and Technology College (permit no. DKY2021021).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We thank the AFWS breeding farm of Chifeng for providing samples.
The work was supported by the Shandong Province Nataral Science Foundation (grant no. ZR2020MC167), the Earmarked Fund for Modern China Wool & Cashmere Technology Research System (grant no. CARS-39-05), the Project of Shandong Province Agricultural Variety Program (grant no. 2019LZGC012), and the Central Guidance on Local Science and Technology Development Fund (grant no. YDZX2021082).
This paper was edited by Henry Reyer and reviewed by Thais Correia Costa and one anonymous referee.
An, H. X., Beckmann, M. W., Reifenberger, G., Bender, H. G., and Niederacher, D.: Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation, Am. J. Pathol., 154, 113–118, https://doi.org/10.1016/s0002-9440(10)65257-1, 1999.
Arner, P. and Kulyté, A.: MicroRNA regulatory networks in human adipose tissue and obesity, Nat. Rev. Endocrinol., 11, 276–288, https://doi.org/10.1038/nrendo.2015.25, 2015.
Azad, T., Tashakor, A., and Hosseinkhani, S.: Split-luciferase complementary assay: applications, recent developments, and future perspectives, Anal. Bioanal. Chem., 406, 5541–5560, https://doi.org/10.1007/s00216-014-7980-8, 2014.
Barak, Y., Nelson, M. C., Ong, E. S., Jones, Y. Z., Ruiz-Lozano, P., Chien, K. R., Koder, A., and Evans, R. M.: PPAR gamma is required for placental, cardiac, and adipose tissue development, Mol. Cell, 4, 585–595, https://doi.org/10.1016/s1097-2765(00)80209-9, 1999.
Blanchet, E., Annicotte, J. S., Lagarrigue, S., Aguilar, V., Clapé, C., Chavey, C., Fritz, V., Casas, F., Apparailly, F., Auwerx, J., and Fajas, L.: E2F transcription factor-1 regulates oxidative metabolism, Nat. Cell Biol., 13, 1146–1152, https://doi.org/10.1038/ncb2309, 2011.
Cao, H., Zhang, S., Shan, S., Sun, C., Li, Y., Wang, H., Yu, S., Liu, Y., Guo, F., Zhai, Q., Wang, Y. C., Jiang, J., Wang, H., Yan, J., Liu, W., and Ying, H.: Ligand-dependent corepressor (LCoR) represses the transcription factor C/EBPβ during early adipocyte differentiation, J. Biol. Chem., 292, 18973–18987, https://doi.org/10.1074/jbc.M117.793984, 2017.
Chen, C., Zong, Y., Tang, J., Ke, R., Lv, L., Wu, M., and Lu, J.: miR-369-3p serves as prognostic factor and regulates cancer progression of hepatocellular carcinoma, Per. Med., 18, 375–388, https://doi.org/10.2217/pme-2020-0012, 2021.
Galleggiante, V., De Santis, S., Liso, M., Verna, G., Sommella, E., Mastronardi, M., Campiglia, P., Chieppa, M., and Serino, G.: Quercetin-Induced miR-369-3p Suppresses Chronic Inflammatory Response Targeting C/EBP-β, Mol. Nutr. Food Res., 63, e1801390, https://doi.org/10.1002/mnfr.201801390, 2019.
Gao, H., Bo, Z., Wang, Q., Luo, L., Zhu, H., and Ren, Y.: Salvanic acid B inhibits myocardial fibrosis through regulating TGF-β1/Smad signaling pathway, Biomed. Pharmacother., 110, 685–691, https://doi.org/10.1016/j.biopha.2018.11.098, 2019.
Guo, L., Li, X., and Tang, Q. Q.: Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) β, J. Biol. Chem., 290, 755–761, https://doi.org/10.1074/jbc.R114.619957, 2015.
Han, F., Li, J., Zhao, R., Liu, L., Li, L., Li, Q., He, J., and Liu, N.: Identification and co-expression analysis of long noncoding RNAs and mRNAs involved in the deposition of intramuscular fat in Aohan fine-wool sheep, BMC Genomics, 22, 98, https://doi.org/10.1186/s12864-021-07385-9, 2021a.
Han, F., Zhou, L., Zhao, L., Wang, L., Liu, L., Li, H., Qiu, J., He, J., and Liu, N.: Identification of miRNA in Sheep Intramuscular Fat and the Role of miR-193a-5p in Proliferation and Differentiation of 3T3-L1, Front. Genet., 12, 633295, https://doi.org/10.3389/fgene.2021.633295, 2021b.
Hawkins, P., Prescott, M. J., Carbone, L., Dennison, N., Johnson, C., Makowska, I. J., Marquardt, N., Readman, G., Weary, D. M., and Golledge, H. D.: A Good Death? Report of the Second Newcastle Meeting on Laboratory Animal Euthanasia, Animals (Basel), 6, 50, https://doi.org/10.3390/ani6090050, 2016.
Hu, B., Zhang, Y., Deng, T., Gu, J., Liu, J., Yang, H., Xu, Y., Yan, Y., Yang, F., Zhang, H., Jin, Y., and Zhou, J.: PDPK1 regulates autophagosome biogenesis by binding to PIK3C3, Autophagy, 17, 2166–2183, https://doi.org/10.1080/15548627.2020.1817279, 2021.
Iorio, M. V., Ferracin, M., Liu, C. G., Veronese, A., Spizzo, R., Sabbioni, S., Magri, E., Pedriali, M., Fabbri, M., Campiglio, M., Ménard, S., Palazzo, J. P., Rosenberg, A., Musiani, P., Volinia, S., Nenci, I., Calin, G. A., Querzoli, P., Negrini, M., and Croce, C. M.: MicroRNA gene expression deregulation in human breast cancer, Cancer Res., 65, 7065–7070, https://doi.org/10.1158/0008-5472.Can-05-1783, 2005.
Jia, W., Di, C., and Shi, L.: Applications of lipidomics in goat meat products: Biomarkers, structure, nutrition interface and future perspectives, J. Proteomics, 270, 104753, https://doi.org/10.1016/j.jprot.2022.104753, 2023.
Karkeni, E., Astier, J., Tourniaire, F., El Abed, M., Romier, B., Gouranton, E., Wan, L., Borel, P., Salles, J., Walrand, S., Ye, J., and Landrier, J. F.: Obesity-associated Inflammation Induces microRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function, J. Clin. Endocr. Metab., 101, 1615–1626, https://doi.org/10.1210/jc.2015-3410, 2016.
Kim, H. J., Cho, H., Alexander, R., Patterson, H. C., Gu, M., Lo, K. A., Xu, D., Goh, V. J., Nguyen, L. N., Chai, X., Huang, C. X., Kovalik, J. P., Ghosh, S., Trajkovski, M., Silver, D. L., Lodish, H., and Sun, L.: MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes, Diabetes, 63, 4045–4056, https://doi.org/10.2337/db14-0466, 2014.
Kim, K. H. and Lee, M. S.: Autophagy – a key player in cellular and body metabolism, Nat. Rev. Endocrinol., 10, 322–337, https://doi.org/10.1038/nrendo.2014.35, 2014.
Krek, W. and Nigg, E. A.: Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites, Embo J., 10, 305–316, https://doi.org/10.1002/j.1460-2075.1991.tb07951.x, 1991.
Lagarrigue, S., Lopez-Mejia, I. C., Denechaud, P. D., Escoté, X., Castillo-Armengol, J., Jimenez, V., Chavey, C., Giralt, A., Lai, Q., Zhang, L., Martinez-Carreres, L., Delacuisine, B., Annicotte, J. S., Blanchet, E., Huré, S., Abella, A., Tinahones, F. J., Vendrell, J., Dubus, P., Bosch, F., Kahn, C. R., and Fajas, L.: CDK4 is an essential insulin effector in adipocytes, J. Clin. Invest., 126, 335–348, https://doi.org/10.1172/jci81480, 2016.
Li, B., Huang, X., Yang, C., Ge, T., Zhao, L., Zhang, X., Tian, L., and Zhang, E.: miR-27a Regulates Sheep Adipocyte Differentiation by Targeting CPT1B Gene, Animals (Basel), 12, 28, https://doi.org/10.3390/ani12010028, 2021.
Li, P., Dong, M., and Wang, Z.: Downregulation of TSPAN13 by miR-369-3p inhibits cell proliferation in papillary thyroid cancer (PTC), Bosnian J. Basic Med., 19, 146–154, https://doi.org/10.17305/bjbms.2018.2865, 2019.
Lie, S., Morrison, J. L., Williams-Wyss, O., Suter, C. M., Humphreys, D. T., Ozanne, S. E., Zhang, S., MacLaughlin, S. M., Kleemann, D. O., Walker, S. K., Roberts, C. T., and McMillen, I. C.: Impact of maternal undernutrition around the time of conception on factors regulating hepatic lipid metabolism and microRNAs in singleton and twin fetuses, Am. J. Physiol. Endocrinol. Metab., 310, E148–E159, https://doi.org/10.1152/ajpendo.00600.2014, 2016.
Linhart, H. G., Ishimura-Oka, K., DeMayo, F., Kibe, T., Repka, D., Poindexter, B., Bick, R. J., and Darlington, G. J.: C/EBPalpha is required for differentiation of white, but not brown, adipose tissue, P. Natl. Acad. Sci. USA, 98, 12532–12537, https://doi.org/10.1073/pnas.211416898, 2001.
Liu, H., Li, B., Qiao, L., Liu, J., Ren, D., and Liu, W.: miR-340-5p inhibits sheep adipocyte differentiation by targeting ATF7, Anim. Sci. J., 91, e13462, https://doi.org/10.1111/asj.13462, 2020.
Liu, P., Ma, C., Wu, Q., Zhang, W., Wang, C., Yuan, L., and Xi, X.: MiR-369-3p participates in endometrioid adenocarcinoma via the regulation of autophagy, Cancer Cell Int., 19, 178, https://doi.org/10.1186/s12935-019-0897-8, 2019.
Livak, K. J. and Schmittgen, T. D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods, 25, 402–408, https://doi.org/10.1006/meth.2001.1262, 2001.
Lorente-Cebrián, S., Mejhert, N., Kulyté, A., Laurencikiene, J., Åström, G., Hedén, P., Rydén, M., and Arner, P.: MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-α, PLoS One, 9, e86800, https://doi.org/10.1371/journal.pone.0086800, 2014.
Lou, J., Huang, J., Dai, X., Xie, Y., Dong, M., and Chen, B.: Knockdown of tetraspanin 13 inhibits proliferation of colorectal cancer cells, Int. J. Clin. Exp. Med., 10, 6387–6395, 2017.
Mizushima, N. and Levine, B.: Autophagy in mammalian development and differentiation, Nat. Cell Biol., 12, 823–830, https://doi.org/10.1038/ncb0910-823, 2010.
Mortimer, S. I., Hatcher, S., Fogarty, N. M., van der Werf, J. H. J., Brown, D. J., Swan, A. A., Jacob, R. H., Geesink, G. H., Hopkins, D. L., Edwards, J. E. H., Ponnampalam, E. N., Warner, R. D., Pearce, K. L., and Pethick, D. W.: Genetic correlations between wool traits and meat quality traits in Merino sheep, J. Anim. Sci., 95, 4260–4273, https://doi.org/10.2527/jas2017.1628, 2017.
Moxnes, J. F., Haux, J., and Hausken, K.: The dynamics of cell proliferation, Med. Hypotheses, 62, 556–563, https://doi.org/10.1016/j.mehy.2003.12.007, 2004.
Raj, A. B., Leach, M. C., and Morton, D. B.: Carbon dioxide for euthanasia of laboratory animals, Comp. Med., 54, 470–471, 2004.
Retzer-Lidl, M., Schmid, R. M., and Schneider, G.: Inhibition of CDK4 impairs proliferation of pancreatic cancer cells and sensitizes towards TRAIL-induced apoptosis via downregulation of survivin, Int. J. Cancer, 121, 66–75, https://doi.org/10.1002/ijc.22619, 2007.
Rodriguez-Puebla, M. L., Miliani de Marval, P. L., LaCava, M., Moons, D. S., Kiyokawa, H., and Conti, C. J.: Cdk4 deficiency inhibits skin tumor development but does not affect normal keratinocyte proliferation, Am. J. Pathol., 161, 405–411, https://doi.org/10.1016/s0002-9440(10)64196-x, 2002.
Rosen, E. D., Sarraf, P., Troy, A. E., Bradwin, G., Moore, K., Milstone, D. S., Spiegelman, B. M., and Mortensen, R. M.: PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro, Mol. Cell, 4, 611–617, https://doi.org/10.1016/s1097-2765(00)80211-7, 1999.
Scalavino, V., Liso, M., Cavalcanti, E., Gigante, I., Lippolis, A., Mastronardi, M., Chieppa, M., and Serino, G.: miR-369-3p modulates inducible nitric oxide synthase and is involved in regulation of chronic inflammatory response, Sci. Rep., 10, 15942, https://doi.org/10.1038/s41598-020-72991-8, 2020.
Xu, Q. and Liu, K.: MiR-369-3p inhibits tumorigenesis of hepatocellular carcinoma by binding to PAX6, J. Biol. Reg. Homeos. Ag., 34, 917–926, https://doi.org/10.23812/20-187-a-35, 2020.
Xu, Q., Mariman, E. C. M., Roumans, N. J. T., Vink, R. G., Goossens, G. H., Blaak, E. E., and Jocken, J. W. E.: Adipose tissue autophagy related gene expression is associated with glucometabolic status in human obesity, Adipocyte, 7, 12–19, https://doi.org/10.1080/21623945.2017.1394537, 2018.
Yekta, S., Shih, I. H., and Bartel, D. P.: MicroRNA-directed cleavage of HOXB8 mRNA, Science, 304, 594–596, https://doi.org/10.1126/science.1097434, 2004.
Yin, Y., Yuan, H., Wang, C., Pattabiraman, N., Rao, M., Pestell, R. G., and Glazer, R. I.: 3-phosphoinositide-dependent protein kinase-1 activates the peroxisome proliferator-activated receptor-gamma and promotes adipocyte differentiation, Mol. Endocrinol., 20, 268–278, https://doi.org/10.1210/me.2005-0197, 2006.