the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Milk-y Way: the impact of single-nucleotide polymorphisms on milk production traits in Kilis dairy goats
Osman Biçer
This study aimed to investigate the impact of single-nucleotide polymorphisms (SNPs) on milk production traits in Kilis dairy goats by analyzing the genotypes of POU1F1, PRLR, β-Lg, GH1, and GH2 genes and their association with lactation milk yield (LMY), lactation length (LL) and average daily milk yield (ADMY). Blood samples were collected from 227 goats, and genotyping was performed using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). The results revealed that the frequencies of the genotypes varied among the genes. The polymorphisms were found to be significantly linked with milk production traits. These findings suggest that SNPs of POU1F1/AluI, PRLR/RsaI and β-Lg/SacII are significantly associated with LMY and that the POU1F1-TC genotype, PRLR-TT genotype and β-Lg-AB genotype are associated with higher LMY and ADMY. Additionally, the POU1F1-TC genotype was found to have a longer LL. However, no significant association was found between the GH1 and GH2 genotypes and LMY, LL and ADMY. Overall, this study provides valuable insights into the genetic factors influencing milk production traits in Kilis dairy goats, which can be utilized for the selection of high-yielding animals in breeding programs.
- Article
(953 KB) - Full-text XML
- BibTeX
- EndNote
The accurate selection of dairy goats with superior milk production traits is essential for breeders and consumers. While traditional selection methods have been used in the past, new approaches, such as the use of genetic markers, have the potential to enhance the precision and efficiency of selection programs. Genetic polymorphisms, in particular, have been closely linked to desired traits of interest, making them a valuable tool for breeders and researchers. Identification and utilization of these genetic markers can improve the overall performance of dairy goats and facilitate the selection of animals with desirable traits for breeding programs. Therefore, continued research on genetic polymorphisms in dairy goats is necessary to improve the selection process and meet the increasing demand for high-quality dairy products.
Goat husbandry in Türkiye plays a pivotal role in the field of animal production (Gül et al., 2020; Gursoy, 2006), with an approximate population of 12 325 000 goats (TUIK, 2022). However, traditional breeding programs are limited with respect to achieving genetic progress, which can take several generations. The use of major genes can accelerate genetic progress, but this requires an understanding of the effect levels of these genes, knowing individual genotypes and developing appropriate mating plans. The Kilis goat has emerged as a breed with a unique genetic profile that reflects its complex ancestry due to historical crossbreeding between the Damascus and Hair goat populations in the region (Gül et al., 2020). The Kilis goat is well adapted to the arid and semiarid regions of Türkiye and is known for its high milk yield and fecundity. Therefore, it holds great potential for milk production and genetic improvement programs in Türkiye.
The POU1F1 (POU class 1 homeobox 1) gene plays a critical role in the growth and development of mammals and is secreted from the anterior pituitary gland. POU1F1 is involved in the regulation of various important physiological processes, including somatic growth, mammary gland development and lactation (Lan et al., 2009; Bastos et al., 2006). Genetic variations in the POU1F1 gene have been found to be associated with variations in milk production traits in dairy goats, making it a promising target for genetic selection programs aimed at improving milk yield and quality (Işık, 2016; Daga et al., 2013; Mura et al., 2012). Prolactin (PRL) is a peptide hormone that plays a crucial role in the development and function of the mammary gland, including milk production and the expression of milk protein genes (Viitala et al., 2006). The biological effects of prolactin are exerted through its interaction with the prolactin receptor (PRLR). The genetic relationship between the PRL and PRLR loci provides valuable insights into the metabolic and functional aspects of the prolactin endocrine axis (Hou et al., 2013). In particular, variations in the PRLR gene have been linked to mammary gland development and differentiation as well as to milk production traits and milk composition in livestock species (Skrzypczak et al., 2015; Hou et al., 2014, 2013; Lü et al., 2011). β-lactoglobulin is a key whey protein found in ruminant milk and belongs to the lipocalin superfamily of hydrophobic molecule carriers. The gene encoding β-lactoglobulin is highly expressed during lactation, particularly in the mammary gland (Mercier and Vilotte, 1993). The expression of this gene is tightly regulated by a complex hormonal mechanism, with prolactin being the main lactogenic stimulus. The β-Lg gene has been found to have polymorphisms that are particularly associated with various milk production traits, such as milk quality, yield and composition, in different ruminant species (Yousefi et al., 2013; Erdoğan, 2010). The growth hormone (GH) gene is part of a large gene family that includes prolactin (PRL) and placental lactogens (CS, chorionic somatomammotropins) (Harvey et al., 1995). GH is specifically expressed from somatotrophic cells in the anterior pituitary gland, and its transcription is regulated by cell-type-specific hormonal mechanisms. Therefore, GH serves as a notable model for understanding the cell type-specific control mechanisms of gene expression during organ development. As the name implies, GH is a crucial hormone for growth and development, and it is also involved in the processes of sexual differentiation and pubertal maturation. GH also participates in gonadal steroidogenesis, gametogenesis and ovulation, and it influences pregnancy and lactation (Hull and Harvey, 2001). Thus, a better understanding of the functional significance of the these genes in relation to milk production traits can provide valuable insights for the development of effective breeding strategies in dairy goat farming.
This study aimed to investigate polymorphisms in the POU1F1, PRLR, β-Lg and GH genes in Kilis goats as well as their relationships with lactation milk yield, lactation length and average lactation milk yield. These polymorphisms can serve as useful genetic markers for animal breeding via marker-assisted selection (MAS).
2.1 Animal selection, data collection and DNA sampling procedure
The Kilis and Damascus (Shami) goat breeds are renowned for their notable milk production and reproductive attributes. These breeds are primarily reared in the provinces of Kilis, Gaziantep, Hatay and Şanlıurfa, situated in the southern region of Türkiye.
It is thought that Kilis goats originated from the uncontrolled mating of Damascus (Shami) goats from Syria and Hair goats, which are one of the domestic goat breeds in Türkiye, under conventional breeding conditions (Gül et al., 2020; Keskin et al., 2017). The unrelated animal subjects of the study comprised 227 Kilis goats, which were obtained from the National Breeding Project of Kilis Goats under Farm Conditions. The project was supported by the General Directory of Agricultural Research and Policy of the Republic of Türkiye’s Ministry of Agriculture and Forestry in Kilis Province. All goats were aged between 2 and 4 years and raised under uniform management and feeding conditions. Individual and total milk yields were recorded every 28 d until the end of lactation. Blood samples were collected from the vena jugularis of each dairy goat using vacuum tubes containing anticoagulant ethylenediaminetetraacetic acid (BD Vacutainer Systems, Plymouth, UK). Approximately 9 mL of blood was collected from each animal. The genomic DNA was extracted from white blood cells using the high-salt method procedure (Miller et al., 1988) and was stored at −20 ∘C until use.
2.2 Polymerase chain reaction (PCR) reactions and genotyping
In this study, two pairs of primers were designed to amplify the goat POU1F1, PRLR, β-Lg, GH1 and GH2 genes. Table 1 provides detailed information on the primer sequences and their corresponding products.
Table 1The primer sequences and information on the genes.
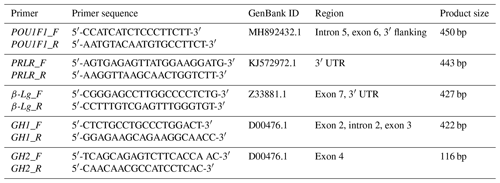
UTR denotes untranslated regions and “bp” stands for base pairs.
The PCR reactions were carried out using genomic DNA and specific concentrations of reagents (Table 2). The thermal cycling protocols involved denaturation, annealing and extension steps, which varied with respect to temperature and time for each gene. The PCR amplification was performed using the C1000 Touch™ thermal cycler (Bio-Rad, USA). To analyze the PCR products of the POU1F1, PRLR, β-Lg and GH genes, aliquots of 30 µL were digested with 10 units of restriction enzymes AluI, RsaI, SacII and HaeIII (Thermo Scientific, USA) for 2 h at 37 ∘C, according to the supplier's instructions. The resulting digested products were then visualized by electrophoresis in a 2.0 % agarose gel stained with ethidium bromide.
Table 2PCR reactions and the cycling protocol.
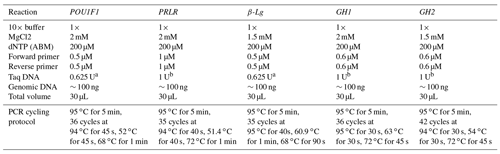
a The Taq DNA polymerase enzyme and buffer from New England Biolabs (NEB), with the amount utilized expressed as units (U), were used. b The Taq DNA polymerase enzyme and buffer from Thermo Scientific USA, with the amount utilized expressed as units (U), were used.
2.3 Statistical analysis
In this study, the goats utilized were granted unrestricted access to water. The lactation milk yield was determined through regular milk control measures taken every 28 d. The AT method from the International Committee for Animal Recording (ICAR) was employed to estimate the lactation milk yield, utilizing the following equation:
where MY denotes the milk yield of goats on the control day, IMY refers to the individual milk yield of each goat during morning milking, TMF indicates the total milk yield of all animals during morning and evening milking, and TF represents the total milk yield of all animals during morning milking. In previous studies, Gül et al. (2016) employed the Fleishman method to calculate the milk yield of each goat.
To establish a relationship between animal genotypes and lactation milk yield, the general linear model of SPSS (Kinnear and Gray, 1999) was used, with lactation length and average daily lactation milk yield serving as parameters. The model was as follows:
where Yij represents the trait measured on the ijth animal, μ refers to population means, αi denotes the effect of the ith genotype and eij represents the effect of random error.
The experimental cohort comprised animals that had given birth either twice or three times. When calculating milk yields, values were standardized based on the overall average. Additive correction factors were applied to reduce variations caused by differences in the number of births, birth type, and farms
Allelic frequencies, genotyping frequencies and Hardy–Weinberg equilibriums were calculated using the GenAlEx 6.5 (Peakall and Smouse, 2012) and POPGENE 32 (Yeh et al., 2000) software packages.
3.1 Genotyping
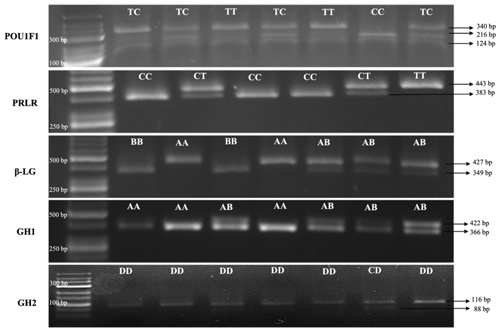
Electrophoretic analysis of DNA patterns resulting from AluI endonuclease restriction of the goat POU1F1 gene (using the GeneRuler Low Range DNA Ladder from Thermo Scientific) revealed three distinct genotypes: CC, TT, and TC. The CC genotype exhibited fragments of sizes 216, 124 and 110 bp. The TT genotype displayed fragments of sizes 340 and 110 bp, while the TC genotype manifested fragments of sizes 340, 216, 124 and 110 bp (Lan et al., 2007). Due to challenges associated with the precise resolution of the 110 bp fragment on a 2 % agarose gel, it was observed that genotypes CC and TC presented with two to three fragments that were imperceptible under visual inspection. Notably, the fragments spanning 340 and 216 bp demonstrated robust visibility and efficacy in accurately delineating the genotypic variations.
After digestion with RsaI endonuclease of the goat PRLR gene (GeneRuler 50 bp DNA Ladder, Thermo Scientific), the CC (383 and 60), CT (443, 383 and 60) and TT (443) genotypes were obtained as a result of the reaction of PCR products with the restriction endonuclease enzyme (Hou et al., 2014). Due to challenges associated with the precise resolution of the 60 bp fragment on a 2 % agarose gel, it was observed that genotypes CC and CT presented with one or two fragments that were imperceptible under visual inspection. Notably, the fragments spanning 443 and 383 bp demonstrated robust visibility and efficacy in accurately delineating the genotypic variations.
After digestion with SacII endonuclease of the goat β-Lg gene (GeneRuler 50 bp DNA Ladder, Thermo Scientific), the AA (427), AB (427, 349 and 78) and BB (349 and 78) genotypes were obtained as a result of the reaction of PCR products with restriction endonuclease enzyme (Pena et al., 2000). Due to challenges associated with the precise resolution of the 78 bp fragment on a 2 % agarose gel, it was observed that genotypes AB and BB presented with one or two fragments that were imperceptible under visual inspection. Notably, the fragments spanning 427 and 349 bp demonstrated robust visibility and efficacy in accurately delineating the genotypic variations.
After digestion with HaeIII endonuclease of the goat GH1 gene (GeneRuler 50 bp DNA Ladder, Thermo Scientific), the AA (366 and 56), AB (422, 366 and 56) and BB (422) genotypes were obtained as a result of the reaction of PCR products with restriction endonuclease enzyme (Hua et al., 2009). Due to challenges associated with the precise resolution of the 76 bp fragment on a 2 % agarose gel, it was observed that genotypes AA and AB presented with two or three fragments that were imperceptible under visual inspection. Notably, the fragments spanning 422 and 366 bp demonstrated robust visibility and efficacy in accurately delineating the genotypic variations. The BB genotype was not found in Kilis goats in this study.
After digestion with HaeIII endonuclease of the goat GH2 gene (GeneRuler Low Range DNA Ladder, Thermo Scientific), the CC (88 and 28), CD (116, 88 and 28) and DD (116) genotypes were obtained as a result of the reaction of PCR products with restriction endonuclease cenzyme (Hua et al., 2009). Due to challenges associated with the precise resolution of the 28 bp fragment on a 2 % agarose gel, it was observed that the genotype CD presented with one fragment that was imperceptible under visual inspection. Notably, the fragments spanning 116 and 88 bp demonstrated robust visibility and efficacy in accurately delineating the genotypic variations. The CC genotype was not found in Kilis goats in this study.
Figure 1 illustrates the fragments of DNA that have undergone cleavage by restriction enzymes in the study.
3.2 Allelic and genotypic frequencies of the genes
The present study utilized the polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) method to detect allelic and genotypic frequencies of various genes, including POU1F1/AluI, PRLR/RsaI, β-Lg/SacII, GH1/HaeIII and GH2/HaeIII, in caprine genomic DNA. The results are presented in Table 3, where the frequencies of different alleles for each locus were determined.
Table 3Allelic and genotyping frequencies of the genes.
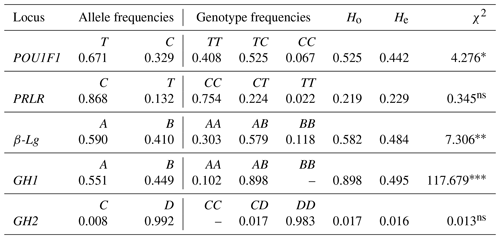
Observed (Ho) and expected (He) heterozygosity and the chi-square (χ2) statistic. The following levels of significance are utilized: * P<0.05, P<0.01, P<0.001 and non-significant (ns).
For the POU1F1/AluI locus, the T and C allele frequencies were 0.671 and 0.329, respectively. The C and T allele frequencies for the PRLR/RsaI locus were 0.868 and 0.132, respectively. Similarly, the A and B allele frequencies for the β-Lg/SacII locus were 0.590 and 0.410, respectively. For the GH1/HaeIII locus, the A and B allele frequencies were 0.551 and 0.449, respectively, and no BB genotype was observed. For the GH2/HaeIII locus, the C and D allele frequencies were 0.008 and 0.992, respectively, and no CC genotype was observed. Furthermore, all genotype distributions for the loci mentioned above were found to be at Hardy–Weinberg equilibrium, except for POU1F1/AluI (P<0.05), β-Lg/SacII (P<0.01) and GH1/HaeIII (P<0.001).
The method from Nei et al. (1983) was used to determine the observed heterozygosity (Ho) and expected heterozygosity (He) values for each locus. For the POU1F1-AluI locus, the Ho and He values were 0.525 and 0.442, respectively. Similarly, for the PRLR/RsaI locus, the Ho and He values were 0.219 and 0.229, respectively. The Ho and He values for the β-Lg/SacII locus were 0.582 and 0.484, respectively. For the GH1/HaeIII locus, the Ho and He values were 0.898 and 0.495, respectively. Lastly, for the GH2/HaeIII locus, the Ho and He values were 0.017 and 0.016, respectively.
3.3 Genetic variations and milk performance in goat populations
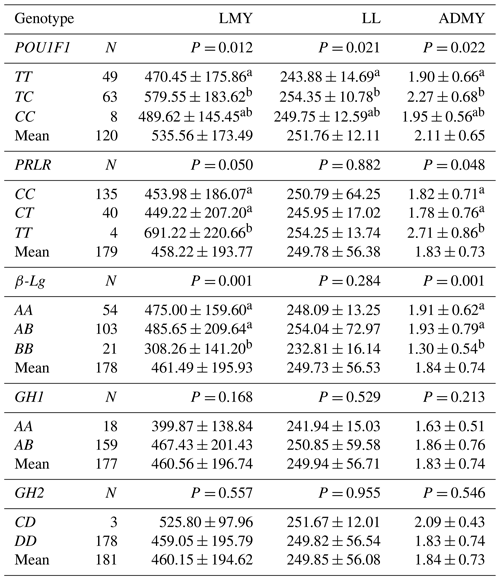
Table 4 presents the effects of the POU1F1, PRLR, β-Lg, GH1 and GH2 genotypes on lactation traits in Kilis goats. The results showed that individuals with the TC genotype in the POU1F1 locus had a considerably (P=0.012) higher lactation milk yield than those with the TT genotype (579.55 ± 183.62 kg vs. 470.45 ± 175.86 kg, respectively), but there was no difference between TT vs. CC or TC vs. CC. Similarly, animals with the TC genotype had a significantly (P=0.021) longer lactation length than animals with the TT genotype (254.35 ± 10.78 d vs. 243.88 ± 14.69 d, respectively), but there was no difference between TT vs. CC or TC vs. CC. Furthermore, Kilis goats with the TC genotype had a significantly (P=0.022) higher average daily milk yield than animals with the TT genotype (2.27 ± 0.68 kg vs. 1.90 ± 0.66 kg, respectively), but there was no difference between TT vs. CC or TC vs. CC.
Regarding the PRLR locus, individuals with the TT genotype had a significantly (P=0.050) greater lactation milk yield than those with the CC and CT genotypes (691.22 ± 220.66 vs. 453.98 ± 186 and 449.22 ± 207.20 kg, respectively). The same pattern was observed for the average daily milk yield (2.71 ± 0.86 vs. 1.82 ± 0.71 and 1.78 ± 0.76 kg, respectively). There was no significant difference between the TT, TC and CC genotypes for lactation length; however, Kilis goats with the TT genotype generally had longer lactation length than animals with the TC and CC genotypes.
With respect to the β-Lg locus, individuals with the AB genotype had a significantly (P=0.001) higher lactation milk yield than those with the BB genotypes (485.65 ± 209.64 vs. 308.26 ± 141.20 kg, respectively), but there was no difference between the AA and AB genotypes. The same pattern was observed for the average daily milk yield (1.93 ± 0.79 vs. 1.30 ± 0.54, respectively). There was no significant difference between the AA, AB and BB genotypes for lactation length; however, animals with the AB genotype generally had longer lactation length than those with AA and BB genotypes.
The AB genotype in the GH1 locus showed a higher lactation milk yield, lactation length and average daily milk yield than the AA genotype, but the differences were not statistically significant. Moreover, the CD genotype in the GH2 locus exhibited a higher lactation milk yield, lactation length and average daily milk yield than the DD genotype, but the differences were statistically insignificant.
In the current study, the TC genotype of POU1F1 demonstrated superior values across all examined traits, whereas the TT genotype exhibited the lowest values. The differences observed among genotypes were statistically significant for the LMY (P = 0.012), LL (P = 0.021) and ADMY (P = 0.022) in Kilis goats. Our findings align with previous research, which has also reported a greater LMY in goats with the TC genotype (Işık, 2016; Lan et al., 2009, 2007). Previous studies have also reported a relationship between different parts of the POU1F1 gene and milk yield in various animal species, including goats, sheep and cattle. Zhou et al. (2016) found an association between the POU1F1/AluI locus and the LMY, while other authors have reported associations between different parts of the POU1F1 gene and the LMY (Özmen et al., 2014; Lan et al., 2013; Mura et al., 2012) and milk production traits in cattle (Khaizaran and Al-Razem, 2014). However, in the study conducted by Huang et al. (2008), a significant effect of the POU1F1 gene on the LMY was observed only for exon 3, whereas no effect was observed for intron 5 nor exon 6 (Trakovicka et al., 2014; Yan et al., 2011). In our study, the TC genotype of POU1F1 was associated with a longer lactation length compared with the CC and TT genotypes. Although studies on lactation period are limited, some other studies have also reported significant effects of the POU1F1 gene on lactation length in sheep (Al-Khuzai and Al-Anbari, 2018) and cattle (Edriss et al., 2009). Moreover, we found a significant relationship between genotypes and the ADMY, with the TC genotype showing a higher ADMY than the other genotypes.
The TT genotype of PRLR displayed the highest value for all evaluated traits. Significant differences were observed among the genotypes with respect to the LMY (P = 0.050) and ADMY (P = 0.048) but not the LL in Kilis goats. Although the differences between the CC and CT genotypes were not statistically significant for LL, our results indicate that the TT genotype has a positive effect on the LL. Recently, most studies have focused on the association of the PRLR gene with reproductive traits in different species, particularly in pigs. In their investigation, El-Shorbagy et al. (2022) underscored that individuals exhibiting the CT genotype displayed a notably elevated daily average milk yield as well as overall milk yield in comparison with those possessing the TT genotype in Egyptian Zaraibi goats. Hou et al. (2014) reported that the PRLR/RsaI locus had an impact on the LMY, but the TT genotype was not observed in the studied populations. In addition, they found that the CC genotype was associated with a higher lactation milk yield. However, numerous studies have reported associations between different genotypes from various regions of the PRLR gene and milk production traits in different species, including the promoter region of PRLR (Fontanesi et al., 2014), intron 2 and exon 9 (Hou et al., 2013), and exon 10 (Lü et al., 2011) in goats as well as exon 3 and exon 7 (Zhang et al., 2008) in cattle. To our knowledge, no studies have investigated the impact of PRLR genotypes on the LL and ADMY in Kilis goats or other species. Therefore, further studies are required to confirm our findings and to explore the role of PRLR genotypes in lactation performance across different breeds and species.
Our findings in Table 4 also reveal an association between genotypes and the tested milk production traits. Specifically, we observed that individuals with the AB genotype of β-Lg had the highest values of LMY, LL and ADMY. While the differences between the AA and AB genotypes were not statistically significant for the LL, we found a significant association between the AB genotype and the LMY (P = 0.001) as well as the ADMY (P = 0.001). Although the differences in the LL were not statistically significant, we did observe a positive effect of the AA and AB genotypes on the LL. This is in agreement with previous studies that also reported an effect of the β-Lg gene on the LMY in goats (Dettori et al., 2015). For instance, Kahilo et al. (2014) reported that individuals with the AB genotype had a greater LMY in Nubian goats, which is consistent with our findings. Similarly, other studies have reported that the AA genotype had a higher LMY than other genotypes in certain goat populations (El-Hanafy et al., 2015, 2010; Kahilo et al., 2014). However, there have also been studies showing no significant effect of the β-Lg gene on the LMY in Saanen and Pakistani domestic goat breeds (Işık et al., 2017; Mahmood et al., 2016). Similarly, in their study, Wardani et al. (2022) noted that there was no statistically significant difference in the association between milk yield and β-lactoglobulin in Senduro goats. Conversely, findings by Dokic et al. (2020) demonstrated a correlation between β-lactoglobulin (β-Lg) polymorphism and milk performance in sheep. The study revealed the prevalence of the BB genotype over the other variants (AA and AB) in terms of milk yield, while no statistically significant disparities among these genotypes were observed across different sheep breeds. The research conducted by Wafaa et al. (2019) unveiled noteworthy distinctions in overall milk production; specifically, the genetic structures denoted as AA exhibited superiority over the genetic structures marked as AB, with a difference in the kilograms of total milk yield. However, no significant variances were observed between the two genetic structures concerning lactation duration. In Çine Çapari sheep, it has been reported that individuals with the AA genotype have a longer LL compared with other genotypes, but no similar studies have been conducted on goats in terms of the relevant locus (Erdoğan, 2010). Additionally, studies on the β-Lg gene in other regions have reported significant effects of the gene on the ADMY in buffalo (Vohra et al., 2012) but no significant effect on the ADMY in sheep (Triantaphyllopoulos et al., 2017).
Our statistical analysis revealed that there was no significant association between GH1/HaeIII, GH2/HaeIII and milk production traits. This finding is consistent with previous reports that demonstrated no relationship between GH1 and milk yield in Algarvia goats (Malveiro et al., 2001). Likewise, GH/TaqI was also found to be insignificant, and the GH/AluI locus was monomorphic in Kankrej cattle (Falaki et al., 1997). However, individuals with the AB genotype had a higher lactation milk yield in Serrana and Sarda goats (Dettori et al., 2013; Marques et al., 2003) as well as in cattle (Krasnopiorova et al., 2012).
Interestingly, GH2 had a significant effect on the LMY in Algarvia and Serrana goats (Marques et al., 2003; Malveiro et al., 2001), but its effect was found to be insignificant in Sarda goats (Dettori et al., 2013). In our study, although the differences between genotypes were statistically insignificant, individuals with AB and CD genotypes had higher values, suggesting a positive effect on the LMY, LL and ADMY.
Our study suggests that the genetic variations present in the POU1F1/AluI, PRLR/RsaI and β-Lg/SacII loci are associated with milk production traits in dairy goats. This finding suggests that this markers may be useful for improving milk production traits in dairy goat breeding programs. Therefore, our results could provide a valuable reference for implementing MAS in the breeding and genetics of dairy goats, with the aim of improving milk production traits.
The results of our study provide important information for genetic improvement programs in the dairy goat industry. The identification of genetic markers associated with milk production traits can help breeders to select the best animals for breeding purposes, leading to more efficient and profitable dairy production. The use of MAS can facilitate the selection of superior individuals for milk production traits and can accelerate genetic progress in dairy goats. Therefore, our findings have important implications for the development of breeding strategies that focus on improving milk production traits in dairy goats.
However, it is worth noting that our study has some limitations, such as the small sample size and the focus on a single goat breed. Further studies with larger sample sizes and involving multiple goat breeds are needed to validate our findings and to investigate the generalizability of these genetic markers to other goat populations. Nevertheless, our study contributes to the understanding of the genetic basis of milk production traits in dairy goats and provides a basis for future studies in this area.
In accordance with the principles of scientific transparency and reproducibility, the data used and analyzed in this study are available from the corresponding author upon reasonable request.
ZG: study design, development of the methodology, data collection, laboratory and statistical analysis, and writing original draft; OB: supervision.
The contact author has declared that neither of the authors has any competing interests.
All procedures carried out in this study that involved animals were approved by The Animal Experiments Local Ethics Committee of Hatay Mustafa Kemal University, based on document number 2016/8-1.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This article is derived from the author's doctoral thesis at Hatay Mustafa Kemal University. The authors acknowledge the Republic of Türkiye's Ministry of Agriculture and Forestry for providing animal materials from “The National Breeding Project of Kilis Goats under Farm Conditions”, which was backed by the General Directorate of Agricultural Research and Policy. The authors also extend their appreciation to the Adnan Menderes University Agricultural Biotechnology and Food Safety Application and Research Center (ADU-TARBIYOMER) for providing laboratory facilities to conduct molecular genetic analysis.
This study received support from the Hatay Mustafa Kemal University Scientific Research Projects Institutional Coordinator (project no. 16460).
This paper was edited by Henry Reyer and reviewed by Raji Kanakkaparambil, Shaimaa A. Mohamed, and one anonymous referee.
Al-Khuzai, H. M. and Al-Anbari, N. N.: Relationship of POU1F1 gene polymorphism with some of economical traits in Iraqi Awassi ewes, J. Entomol. Zool. Stud., 6, 2082–2085, 2018.
Bastos, E., Santos, I., Parmentier, I., Castrillo, J. L., Cravador A., Guedes-Pinto, H., and Renaville, R.: Ovis aries POU1F1 gene: cloning, characterization and polymorphism analysis, Genetica, 126, 303–314, https://https://doi.org/10.1007/s10709-005-0034-6, 2006.
Daga, C., Paludo, M., Luridiana, S., Mura, M. S., Bodano, S., Pazzola, M., Dettori, M. L., Vacca, G. M., and Carcangiu, V.: Identification of novel SNPs in the Sarda breed goats POU1F1 gene and their association with milk productive performance, Mol. Biol. Rep., 40, 2829–2835, https://https://doi.org/10.1007/s11033-012-2298-0, 2013.
Dettori, M. L., Rocchigiani, A. M., Luridiana, S., Mura, M. C., Carcangiu, V., Pazzola, M., and Vacca, G. M.: Growth hormone gene variability and its effects on milk traits in primiparous Sarda goats, J. Dairy Res., 80, 255–262, https://doi.org/10.1017/S0022029913000174, 2013.
Dettori, M. L., Pazzola, M., Pira, E., Puggioni, O., and Vacca, G. M.: Variability of the caprine whey protein genes and their association with milk yield, composition and renneting properties in the Sarda breed: 2. The BLG gene, J. Dairy Res., 82, 442–448, https://doi.org/10.1017/S0022029915000473, 2015.
Dokic, M., Markovic, B., Gantner, V., and Markovic, M.: Association of genetic variants of β-lactoglobulin gene with milk traits of Jezeropivska sheep breed, Agriculture&Forestry, 66, 15–23, https://doi.org/10.17707/AgricultForest.66.3.02, 2020.
Edriss, M. A., Edriss, V., and Rahmani, H. R.: Association of PIT-1 gene polymorphism with birth weight, milk and reproduction traits in Isfahan Holstein cows, Archiv Tierzucht, 52, 445–447, https://doi.org/10.5194/aab-52-445-2009, 2009.
El-Hanafy, A. A., El-Saadani, M. A., Eissa, M., Maharem, G. M., and Khalifa, Z. A.: Polymorphism of β-lacto Globulin gene in Barki and Damascus and their crossbreed goats in relation to milk yield, Biotechnol. Anim. Husb., 26, 1–12, https://doi.org/10.2298/BAH1002001E, 2010.
El-Hanafy, A. A. M., Qureshi, M. I., Sabir, J., Mutawakil, M., Ahmed, M. M. M., El-Ashmaoui, H., Ramadan, H. A. M. I., Abou-Alsoud, M., and Sadek, M. A.: Nucleotide sequencing and DNA polymorphism studies of beta-lactoglobulin gene in native Saudi goat breeds in relation to milk yield, Czech J. Anim. Sci., 60, 132–138, https://doi.org/10.17221/8078-CJAS, 2015.
El-Shorbagy, H. M., Abdel-Aal, E. S., Mohamed, S. A., and El-Ghor, A. A.: Association of PRLR, IGF1, and LEP genes polymorphism with milk production and litter size in Egyptian Zaraibi goat, Trop. Anim. Health Pro., 54, 321, https://doi.org/10.1007/s11250-022-03316-2, 2022.
Erdoğan, F.: Milk yield characteristics and β-lactoglobulin gene polymorphism in indigenous Çine Çapari sheep, MSc thesis, Thesis no. 259550, Adnan Menderes University, Aydın, Turkey, https://tez.yok.gov.tr/UlusalTezMerkezi/tezSorguSonucYeni.jsp (last access: 23 November 2023), 2010.
Falaki, M., Prandi, A., Corradini, C., Sneyers, M., Gengler, N., Massart, S., Fazzini, U., Burny, A., Portetelle, D., and Renaville, R.: Relationship of growth hormone gene and milk protein polymorphisms to milk production traits in Simmental cattle, J. Dairy Res., 64, 47–56, https://doi.org/10.1017/s0022029996001872, 1997.
Fontanesi, L., Calo, D. G., Galimberti, G., Negrini, R., Marino, R., Nardone, A., Ajmone-Marsan, P., and Russo, V.: A candidate gene association study for nine economically important traits in Italian Holstein cattle, Anim. Genet., 45, 576–580, https://doi.org/10.1111/age.12164, 2014.
Gursoy, O.: Economics and profitability of sheep and goat production in Turkey under new support regimes and market conditions, Small Ruminant. Res., 62, 181–191, https://doi.org/10.1016/j.smallrumres.2005.08.013, 2006.
Gül, S., Keskin, M., Göçmez, Z., and Gündüz, Z.: Effects of supplemental feeding on performance of Kilis goats kept on pasture condition, Ital. J. Anim. Sci., 15, 110–115, https://doi.org/10.1080/1828051X.2015.1132542, 2016.
Gül, S., Yilmaz, O., Gündüz, Z., Keskin, M., Cemal, İ., Ata, N., and Önel, S. E.: The genetic structure of the goat breeds belonging to Northwest part of Fertile Crescent, Small Ruminant. Res., 182, 22–28, https://doi.org/10.1016/j.smallrumres.2019.09.009, 2020.
Harvey, S., Scanes, C. G., and Doughaday, W. H.: Growth Hormone. Basic, Clinical and Applied Aspects, Boca Raton, CRC Press Inc., FL, USA, ISBN 978-0849386978, 1995.
Hou, J. X., An, X. P., Song, Y. X., Wang, J. G., Ma, T., Han, P., Fang, F., and Cao, B. Y.: Combined effects of four SNPs within goat PRLR gene on milk production traits, Gene, 529, 276–281, https://doi.org/10.1016/j.gene.2013.07.057, 2013.
Hou, J. X., Fang, F., An, X. P., Yan, Y., Ma, T., Han, P., Meng, F. X., Song, Y. X., Wang, J. G., and Cao, B. Y.: Polymorphisms of PRLR and FOLR1 genes and association with milk production traits in goats, Genet. Mol. Res., 13, 2555–2562, https://doi.org/10.4238/2014.January.24.1, 2014.
Hua, G. H., Chen, S. L., Yu, J. N., Cai, K. L., Wu, C. J., Li, Q. L., Zhang, C. Y., Liang, A. X., Han, L., Geng, L. Y., Shen, Z., Xu, D. Q., and Yang, L. G.: Polymorphism of the growth hormone gene and its association with growth traits in Boer goat bucks, Meat Sci., 81, 391–395, https://https://doi.org/10.1016/j.meatsci.2008.08.015, 2009.
Huang, W., Maltecca, C., and Khatib, H.: A proline-to-histidine mutation in POU1F1 is associated with production traits in dairy cattle, Anim. Genet., 39, 554–557, https://doi.org/10.1111/j.1365-2052.2008.01749.x, 2008.
Hull, K. L. and Harvey, S.: Growth hormone: Roles in female reproduction, J. Endocrinol., 168, 1–23, https://doi.org/10.1677/joe.0.1680001, 2001.
Işık, R.: Determination of POU1F1 gene polymorphism and its association with production traits in Saanen goats, PhD thesis, Thesis no. 436572, Ege University, İzmir, Turkey, https://tez.yok.gov.tr/UlusalTezMerkezi/tezSorguSonucYeni.jsp (last access: 23 November 2023), 2016.
Işık, R., Bilgen, G., Koşum, N., Kandemir, Ç., and Taşkın, T.: Polymorphism in exon 7 of β-lactoglobulin (β-LG) gene and its association with milk yield in Saanen goats, Journal of Tekirdağ Agricultural Faculty, special issue, 35–40, 2017.
Kahilo, K., El-Shazly, S., El-Khadrawy, A., and Fattouh, I.: Genetic polymorphism in β-lactoglobulin gene of some goat breeds in Egypt and its influence on milk yield, Life Sci. J., 11, 232–238, 2014.
Keskin, M., Gül, S., Biçer, O., and Daşkıran, İ.: Some reproductive, lactation, and kid growth characteristics of Kilis goats under semiintensive conditions, Turk. J. Vet. Anim. Sci., 41, 248–254, https://doi.org/10.3906/vet-1604-33, 2017.
Khaizaran, Z. A. and Al-Razem, F.: Analysis of selected milk traits in Palestinian Holstein-Friesian cattle in relation to genetic polymorphism, J. Cell Anim. Biol., 8, 74–85, https://doi.org/10.5897/JCAB2014.0409, 2014.
Kinnear, P. R. and Gray, C. D.: SPSS for Windows Made Simple, 3rd Edn., Department of Psychology University of Aberdeen, UK, ISBN 0-86377-611-6, 1999.
Krasnopiorova, N., Baltrenaite, L., and Miceikiene, I.: Growth hormone gene polymorphism and its influence on milk traits in cattle breed in Lithuania, Vet. Zootech., 58, 42–46, 2012.
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., Lei, C. Z., Zhang, A. L., and Zhang, L.: An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits, Small Ruminant. Res., 73, 8–12, https://doi.org/10.1016/j.smallrumres.2006.10.009, 2007.
Lan, X. Y., Li, M. J., Chen, H., Zhang, L. Z., Jing, Y. J., Wei, T. B., Ren, G., Wang, X., Fang, X. T., Zhang, C. L., and Lei, C. Z.: Analysis of caprine pituitary specific transcription factor-1 gene polymorphism in indigenous Chinese goats, Mol. Biol. Rep., 36, 705–709, https://doi.org/10.1007/s11033-008-9232-5, 2009.
Lan, X. Y., Zhao, H. Y., Li, Z. J., Zhou, R., Pan, C. Y., Lei, C. Z., and Chen, H.: Exploring the novel genetic variant of PITX1 gene and its effect on milk performance in dairy goats, J. Integr. Agr., 12, 118–126, https://doi.org/10.1016/S2095-3119(13)60212-9, 2013.
Lü, A., Hu, X., Chen, H., Dong, Y., Zhang, Y., and Wang, X.: Novel SNPs of the bovine PRLR gene associated with milk production traits, Biochem. Genet., 49, 177–189, https://doi.org/10.1007/s10528-010-9397-1, 2011.
Mahmood, A., Jamy, M., Saqlain, M., Shaiq, P. A., Naqvi, S. M. S., and Raja, G. K.: Association of β-lactoglobulin protein isoforms with milk constituents in goat breeds, J. Anim. Plant Sci., 26, 862–867, 2016.
Malveiro, E., Pereira, M., Marques, P. X., Santos, I. C., Belo, C., Renaville, R., and Cravador, A.: Polymorphisms at five exons of the growth hormone gene in the Algarvia goat: possible association with milk traits, Small Ruminant. Res., 41, 163–170, https://doi.org/10.1016/s0921-4488(01)00198-5, 2001.
Marques, P. X., Pereira, M., Marques, M. R., Santos, I. C., Belo, C. C., Renaville, R., and Cravador, A.: Association of milk traits with SSCP polymorphisms at the growth hormone gene in the Serrana goat, Small Ruminant. Res., 50, 177–185, https://doi.org/10.1016/S0921-4488(03)00104-4, 2003.
Mercier, J. C. and Vilotte, J. L.: Structure and function of milk protein genes, J. Dairy Sci., 76, 3079–3098, https://doi.org/10.3168/jds.S0022-0302(93)77647-X, 1993.
Miller, S. A., Dykes, D. D., and Polesky, H. F.: A simple salting out procedure for extracting DNA from human nucleated cells, Nucleic Acids Res., 16, 1215, https://doi.org/10.1093/nar/16.3.1215, 1988.
Mura, M. C., Daga, C., Paludo, M., Luridiana, S., Pazzola, M., Bodano, S., Dettori, M. L., Vacca, G. M., and Carcangiu, V.: Analysis of polymorphism within POU1F1 gene in relation to milk production traits in dairy Sarda sheep breed, Mol. Biol. Rep., 39, 6975–6979, https://doi.org/10.1007/s11033-012-1525-z, 2012.
Nei, M., Tajima, F., and Tateno, Y.: Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data, J. Mol. Evol., 19, 153–170, https://doi.org/10.1007/BF02300753, 1983.
Özmen, Ö., Kul, S., and Ünal, E. Ö.: Polymorphism of sheep POU1F1 gene exon 6 and 3′UTR region and their association with milk production traits, Iran. J. Vet. Res., 15, 331–335, 2014.
Peakall, R. and Smouse, P. E.: GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update, Bioinformatics, 28, 2537–2539, 2012.
Pena, R. N., Sanchez, A., and Folch, J. M.: Characterization of genetic polymorphism in the goat β-lactoglobulin gene, J. Dairy Res., 67, 217–224, https://doi.org/10.1017/s0022029900004155, 2000.
Skrzypczak, E., Babicz, M., and Pastwa, M.: Effect of prolactin receptor (PRLR), and beta-casein (CSN2) gene polymorphism on the chemical composition of milk sows, Folia Biol., 63, 135–144, https://doi.org/10.3409/fb63_2.135, 2015.
Trakovicka, A., Moravcikova, N., Gabor, M., and Miluchova, M.: Genetic polymorphism of Pit-1 gene associated with milk production traits in Holstein cattle, Acta Agraria Kaposvariensis, 18, 146–151, 2014.
Triantaphyllopoulos, K. A., Koutsouli, P., Kandris, A., Papachristou, D., Markopoulou, K. E., Mataragka, A., Massouras, T., and Bizelis, I.: Effect of β-lactoglobulin gene polymorphism, lactation stage and breed on milk traits in Chios and Karagouniko sheep breeds, Ann. Anim. Sci., 17, 371–384, https://doi.org/10.1515/aoas-2016-0058, 2017.
TUIK: https://data.tuik.gov.tr/Kategori/GetKategori?p=tarim-111&dil=1 (last access: 14 November 2022), 2022.
Viitala, S., Szyda, J., Blott, S., Schulman, N., Lidauer, M., Maki-Tanila, A., Georges, M., and Vilkki, J.: The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat, and protein production in Finnish Ayrshire dairy cattle, Genetics, 173, 2151–2164, https://doi.org/10.1534/genetics.105.046730, 2006.
Vohra, V., Dayal, S., and Bhattacharya, T. K.: SSCP typing of alpha-lactalbumin and beta-lactoglobulin gene and its association with milk production and constituent traits in Indian riverine buffalo, Ind. J. Anim. Sci., 82, 884–888, 2012.
Wafaa, I., Hassooni, H. A., and Alkhazraji, W. J.: Association of β-lactoglobulin gene polymorphism with milk production and composition in local awassi sheep, Plant Arch., 19, 284–288, 2019.
Wardani, F. E., Palayukan, J., Furqon, A., Suyadi, S., and Susilorini, T. E.: Polymorphism of B-Lactoglobulin (B-Lg) Gene and Its Association with Milk Yield and Milk Composition on Senduro Goats, Iranian Journal of Applied Animal Science, 12, 111–118, 2022.
Yan, L. J., Fang, X. T., Zhang, R. F., Zhang, C. L., and Chen, H.: Analysis of pituitary specific transcription factor-1 gene polymorphism in several indigenous Chinese cattle and crossbred cattle, J. Appl. Anim. Res., 39, 269–274, https://doi.org/10.1080/09712119.2011.607920, 2011.
Yeh, F., Yang, R. C., and Boyle, T.: Popgene Microsoft Windows based freeware for population genetic analysis, https://sites.ualberta.ca/~fyeh/ (last access: 5 February 2020), 2000.
Yousefi, S., Azari, M. A., Zerehdaran, S., Samiee, R., and Khataminejhad, R.: Effect of β-lactoglobulin and K-casein genes polymorphism on milk composition in indigenous Zel sheep, Archiv Tierzucht, 56, 216–224, https://doi.org/10.7482/0003-9438-56-021, 2013.
Zhang, J. L., Zan, L. S., Fang, P., Zhang, F., Shen, G., and Tian, W.: Genetic variation of PRLR gene and association with milk performance traits in dairy cattle, Can. J. Anim. Sci., 88, 33–39, https://doi.org/10.4141/CJAS07052, 2008.
Zhou, F., Yang, Q., Lei, C., Chen, H., and Lan, X.: Relationship between genetic variants of POU1F1, PROP1, IGFBP3 genes and milk performance in Guanzhong dairy goats, Small Ruminant. Res., 140, 40–45, https://doi.org/10.1016/j.smallrumres.2016.05.015, 2016.