the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Overexpression of goat STEAP4 promotes the differentiation of subcutaneous adipocytes
Xin Li
Hao Zhang
Yong Wang
Yanyan Li
Yan Xiong
Ruiwen Li
Jiangjiang Zhu
Yaqiu Lin
Objective: The focus of this study was the six-transmembrane epithelial antigen of the prostate 4 (STEAP4) gene, on the basis of the cloned goat STEAP4 gene sequence. Its molecular and expression characteristics were analyzed, and its influence on the differentiation of goat subcutaneous adipocytes was explored through overexpression. Method: Reverse-transcription PCR (RT-PCR) was used to clone the goat STEAP4 sequence, and online tools were used to analyze the molecular characteristic. Real-time quantitative PCR (qPCR) was used to detect the expression level of STEAP4 in goat tissues and subcutaneous adipocyte differentiation. Liposome transfection, BODIPY, Oil Red O staining, and qPCR were used to explore the effect of overexpression of STEAP4 on adipocyte differentiation. Results: The cloned goat STEAP4 gene sequence was 1388 bp, and the complete coding sequence (CDS) region was 1197 bp, which encoded a total of 398 amino acids. Compared with the predicted sequence (XM_005679300.3), there were three base mutations in the CDS region of goat STEAP4, A188G, T281C, and A507G. Among them, A507G changed the amino acid at position 170 from Ile to Val. Analysis of the physical and chemical properties of the protein showed that STEAP4 was a stable hydrophilic basic protein. STEAP4 gene expression level was highest in goat liver tissue (P<0.01), followed by lung and back subcutaneous adipose tissue. STEAP4 showed different expression levels in goat subcutaneous adipocytes at different times during the induction of differentiation. The expression in the late stage of differentiation was higher than that before differentiation and lowest at 12 h (P<0.01). Overexpression of STEAP4 promoted the accumulation of intracellular lipid droplets; C/EBPβ (CCAAT enhancer binding protein) was extremely significantly up-regulated (P<0.01), and aP2 (fatty acid binding protein) was significantly up-regulated (P<0.05). Conclusion: Overexpression of STEAP4 could promote the differentiation of goat subcutaneous preadipocytes. This study lays the foundation for an in-depth study of the role of STEAP4 in goat lipid deposition.
- Article
(4210 KB) - Full-text XML
- BibTeX
- EndNote
Goats can provide humans with important living resources such as cashmere, milk, and chevon and are an important breed of livestock. The Jianzhou Da'er goat is the second independently selected breed of goat for meat in China, which has the advantages of resistance to rough feeding and strong adaptability. The taste of chevon is susceptible to flavor substances, which are mainly deposited in adipose tissue, and fat traits are important economic traits of livestock (Kong et al., 2021). Therefore, analyzing the mechanism of goat fat tissue deposition is helpful for breeders to regulate lipid accumulation at the molecular level, thereby improving meat quality. Lipid deposition mainly depends on the clonal proliferation and expansion of adipocytes, and a variety of genes and transcription factors participate in these processes (Lefterova and Lazar, 2009).
STEAP (STEAP1) was first discovered in 1999, and because of its high expression in early prostate cancer, it was named the prostate six-transmembrane epithelial antigen (Huber et al., 1999). It has a highly conserved six-span membrane structural domain. Later, the genes of STEAP2, STEAP3, and STEAP4 (six-transmembrane epithelial antigen of the prostate 4), which also have a conserved six-pass transmembrane structure, were classified into the STEAP family. Ohgami et al., (2005, 2006) did a systematic study on the sequence, subcellular localization, tissue expression, and metal reductase activity of the human STEAP family. All four members of this family have six transmembrane domains and carboxy-terminal sequences, which mediate the cell reduction of Fe3+ and promote the absorption of Fe3+ and Cu2+ (Oosterheert et al., 2018). STEAP family genes are widely present in eukaryotes and participate in their life activities, such as cell proliferation, differentiation, apoptosis, and other physiological processes (Qin et al., 2010; Kim et al., 2016; Valenti et al., 2010). STEAP4 (six-transmembrane epithelial antigen of the prostate) is the newest member of the prostate six-transmembrane epithelial antigen family (Moldes et al., 2001). It has the strongest metal reductase activity in the STEAP family and plays an important role in cancer cell proliferation and metastasis (Tanaka et al., 2012; Korkmaz et al., 2005). For example, STEAP4, as a target of miR-1265, was related to focal adhesion kinase (FAK) phosphorylation and epithelial–mesenchymal transition (EMT). It was involved in the metastasis of bladder cancer (Yan et al., 2019). The STEAP4 gene was located on the long arm of human chromosome 7 (Moldes et al., 2001), which was known to be a susceptible area for type 2 diabetes and insulin resistance, and obesity was closely related to the occurrence of these diseases. Studies have found that STEAP4 was highly expressed in the subcutaneous and visceral adipose tissue of obese patients (Ozmen et al., 2016). Functional studies have shown that the STEAP4 gene is highly expressed in human preadipocytes, gradually down-regulated with adipocyte differentiation and maturation. It can promote adipocyte differentiation, maturation, and lipid accumulation, which may be related to the occurrence of obesity (Chen et al., 2010). In addition, studies have found that mutations in fat content were related to obesity-related genes FTO, melanocortin receptor-4 (MCR4), histamine, leptin receptor (LEPR), and STEAP4 (Millon-Frémillon et al., 2008). But the effect of STEAP4 on goat adipocyte differentiation and lipid deposition is unclear.
Therefore, this study took the Jianzhou Da'er goat as the research object, cloned the STEAP4 gene in goat sequence by reverse-transcription PCR (RT-PCR), and analyzed its physical and chemical properties. The expression of STEAP4 was detected by qPCR in multiple goat tissues and adipocytes at different stages of differentiation. Further, the constructed pcDNA-3.1-STEAP4 vector was transfected into subcutaneous adipocytes for overexpression. On a morphological and molecular level, the effect on the expression of subcutaneous adipocytes was obvious. This study will provide a basis for elucidating the biological function of the STEAP4 gene in goat and provide a reference for goat molecular breeding.
2.1 Sample collection
The tissue samples cryopreserved in liquid nitrogen were taken from a 1-year-old Jianzhou Da'er male goat (Sichuan Jianyang Dageda Animal Husbandry Co., Ltd, Sichuan, China) (heart, liver, spleen, lung, kidney, longissimus dorsi, back subcutaneous adipose tissue, and abdominal subcutaneous adipose tissue, n=3) (He et al., 2021), and TRIzol reagent (TaKaRa, Tokyo, Japan) was used to extract the total RNA from the tissue. The concentration and quality of the RNA sample were detected by the Implen NanoPhotometer® N60, 1 µg of qualified RNA was taken, and the reverse-transcription kit (Thermo Fisher Scientific, Waltham, USA) was used to reverse-transcribe it into cDNA and store it at −20∘ for later use.
2.2 Cell culture
The primary subcutaneous adipocytes from longissimus dorsi were isolated and cryopreserved as in previous work (He et al., 2021). Briefly, the subcutaneous adipose tissue was isolated from 7 d old Jianzhou da'er goats (n=3) and digested with collagenase type I (Sigma, St. Louis, MO, USA) at 37 ∘C for 60 min. The goat primary subcutaneous adipocytes were resuscitated, cultured in a 37∘ incubator with 5 % CO2, and were passed when the confluence reached 90 %. When the F3 generation cell confluence reached 80 %, a complete medium containing 50 µmol L−1 oleic acid (Sigma, St. Louis, MO, USA) was added to induce differentiation, and they were induced and collected separately at 0, 24, 48, 72, 96, and 120 h. RNA was extracted and reverse-transcribed to obtain cell temporal cDNA.
2.3 Cloning and analysis of the goat STEAP4 gene
According to the predicted sequence of the goat STEAP4 gene on NCBI (XM_00567900.3), primers were designed using Primer Premier 5.0 software (Table 1) and were synthesized by Sangon Biotech (Shanghai, China). The PCR reaction system included 12.5 µL Primer STAR Max DNA Polymerase (TaKaRa, Tokyo, Japan), 1.0 µL (10 µmol L−1) sense and antisense primers, 1.0 µL goat liver tissue cDNA, and 9.5 µL ddH2O . The PCR amplification program was as follows: 98∘C for 3 min, 94∘C for 30 s, 58∘C for 15 s, and 72∘C for 30 s and 38 cycles. The PCR products were purified and recovered by electrophoresis identification, and the recovered product was ligated with PMD-19T (TaKaRa, Tokyo, Japan) and transformed into DH5α (Tiangen, Beijing, China). The positive colonies were screened by an AMP-resistant solid medium, and the gene sequence was obtained by Sangon Biotech. The molecular characteristics of the obtained gene sequence are analyzed, and the analysis items and tools are shown in Table 2.
2.4 Construction of goat pcDNA-3.1-STEAP4 vector and cell transfection
Kpn I and Not I restriction sites were selected to construct the goat STEAP4 expression vector by double-restriction enzyme digestion. Subcloning primers were designed according to the cloned STEAP4 sequence, S: 5′CGGGGTACCATGGAGAAAATTTCTACAGATGC3′ and A: 5′TATTCTTAGCGGCCGCTCAGGATCCTTTCATG3′. The pcDNA-3.1 plasmid and the subcloned product were each digested with double enzymes, and the fragments were purified. The fragments were ligated with T4 ligase (TaKaRa, Tokyo, Japan) overnight at 16 ∘C. After the enzyme digestion was verified, they were transformed into DH5α to screen positive colonies for sequencing. The bacteria solution with correct sequencing was amplified, and then the plasmid was extracted. F3 generation cells were seeded into a 12-well plate with 4×104 cells per well. The master mix was configured according to the transfection reagent instructions and incubated for 15 min at room temperature. The cells were cultured with oleic acid induction solution after 14 h of transfection. The total RNA was collected after 48 h to obtain cDNA by reverse transcription, and the negative control group (NC) was transfected with empty pcDNA-3.1. Three biological replicates in each group.
2.5 BODIPY and Oil Red O staining
Cells used for staining were cultured in 24-well plates. The transfection system corresponds to 12-well plates halved. The culture medium was discarded after 48 h of induction, washed three times with phosphate-buffered saline (PBS), and fixed with 10 % neutral formaldehyde at room temperature for 30 min. It was then washed with PBS three times, 200 µL BODIPY working solution (BODIPY stock solution: PBS = 1 : 1000) (Thermo Fisher Scientific, Waltham, USA) was added to each well under dark conditions, and the lipid droplets were observed after 30 min. For Oil Red O staining, Oil Red O working solution (Oil Red O storage solution: ddH2O = 3 : 2) (Solarbio, Beijing, China) was added, and the red lipid droplets were observed after dyeing for 30 min. ImageJ was used to quantify the staining results.
2.6 Quantitative polymerase chain reaction (qPCR) detection
According to the cloned goat STEAP4 sequence, specific qPCR primers were designed (Table 1) to detect its expression. qPCR was used to detect the expression of STEAP4 and to detect the expression of adipogenic differentiation marker genes after overexpression of STEAP4. The primer information is shown in Table 1. With UXT as the internal reference gene, the qPCR system included 2×10 µL SYBR® Premix Ex Tax™ (TaKaRa, Tokyo, Japan), 1 µL each of the sense and antisense primers, 1 µL cDNA, and ddH2O filled to 20 µL. The qPCR procedure was as follows: 95∘ predenaturation for 3 min, 95∘ denaturation for 10 s, 60∘ annealing for 10 s, and 72∘ extension for 15 s for 40 cycles.
2.7 Data analysis
The results of qPCR data were processed by the 2−ΔΔCt method, expressed as mean ± standard deviation. SPSS 18.0 one-way analysis of variance (ANOVA) was used to test the significance. The T test was used in multiple comparisons. P<0.05 indicates a significant difference, and P<0.01 indicates a extremely significant difference.
3.1 Cloning and sequence analysis of goat STEAP4
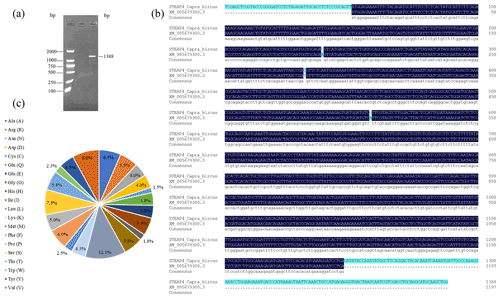
PCR amplification obtained fragments consistent with the expected target product size (Fig. 1a). The length of the goat STEAP4 gene was 1388 bp, including the complete coding sequence (CDS) region of 1197 bp, which was consistent with the predicted sequence (XM_005679300.3). In comparison, there were three base mutations in the CDS region, namely A188G, T281C, and A507G (Fig. 1b). A188G and T281C did not affect protein translation, while A507G changed the amino acid at position 170 from Ile to Val. The STEAP4 gene in goats encodes a total of 398 amino acids, including 48 leucine (L), at the highest proportion (12.1 %), followed by 32 proline (V) (8.0 %) (Fig. 1c).
3.2 Analysis of goat STEAP4 protein characteristics
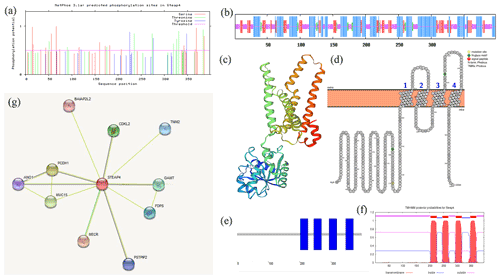
ExPASy was used to analyze the physical and chemical properties of the goat STEAP4 protein. The results showed that the molecular weight of goat STEAP4 protein was 45.327 kD. The total average theoretical isoelectric point, instability index, and hydrophilicity were 9.08, 36.55, and 0.130, respectively, indicating that the goat STEAP4 protein is a stable, hydrophilic basic protein. Since there were more positively charged amino acids than negatively charged amino acids, the goat STEAP4 protein was positively charged. There were 33 potential phosphorylation sites for goat STEAP4 protein, including 17 serine (Ser) phosphorylation sites, 11 threonine (Thr) phosphorylation sites, and 4 tyrosine (Tyr) phosphorylation sites (Fig. 2a). The NetNGlyc tool was used to predict that STEAP4 had two N-glycosylation sites, located at positions 158 and 323, without O glycosylation sites and no signal peptide.
SOPMA was used to predict the secondary structure of STEAP4, and this showed that it contains 42.71 % α helix, 34.17 % random coil, 4.02 % β turn, and 19.10 % stretched chain (Fig. 2b). SWISS-MODEL was used to predict the tertiary structure shown in Fig. 2c, and this was consistent with the results of the secondary-structure prediction.
Using WoLF PSORT to predict subcellular localization showed that STEAP4 was mainly distributed in the endoplasmic reticulum membrane, with a small amount in the cytoplasm. Then, Protter was used to predict the transmembrane structure of STEAP4, showing that STEAP4 crosses the membrane four times through the α helix. And there were two N-glycosylation sites (Fig. 2d), which was consistent with NetNGlyc's prediction results. SMART domain analysis and TMHMM transmembrane structure prediction both showed that goat STEAP4 contains four-transmembrane structures (Fig. 2e, f).
Predicting interacting proteins through the STRING database showed that goat STEAP4 had strong interactions with proteins such as MECR, TNNI2, and CDKL2 (Fig. 2g).
3.3 Conservative analysis of goat STEAP4
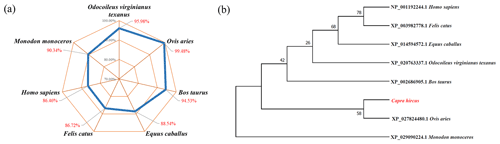
BLAST was used to compare the amino acid similarity of goat STEAP4 with other species. The results showed that the sequence identity of goat STEAP4 and sheep reached 99.48 %, followed by deer and cattle (Fig. 3a). Based on the amino acid sequence of STEAP4, MEGA7.0 was used to construct the phylogenetic tree. The tree showed that goat and sheep were closely related, and cattle and deer were relatively closely related (Fig. 3b).
3.4 Analysis of the goat STEAP4 expression pattern
3.4.1 Tissue distribution of goat STEAP4
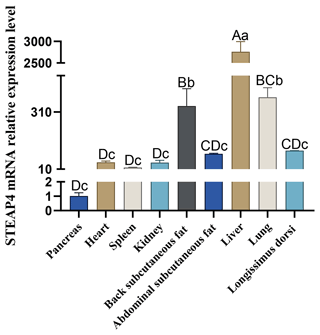
The expression level of STEAP4 in different tissues of goats was detected by qPCR. The expression profile showed that STEAP4 expression was significantly higher in goat liver tissue than other tissues (P<0.01), followed by lungs, also higher in back subcutaneous fat tissues and abdominal subcutaneous fat tissues (Fig. 4).
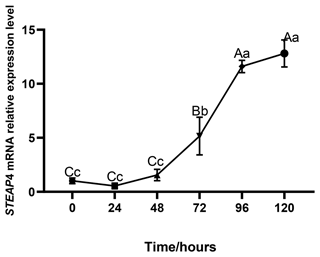
Different capital letters in the figure indicate that the difference is extremely significant (P<0.01), and different lowercase letters indicate significant difference (P<0.05), the same as below.
3.4.2 Cell temporal expression profile of goat STEAP4
The expression level of STEAP4 was expressed at different levels at different times of subcutaneous adipocyte differentiation in goats. The expression of STEAP4 showed a decreasing and then an increasing trend, reaching the lowest level at 24 h after differentiation (Fig. 5).
3.5 Overexpression of goat STEAP4 promotes the differentiation of subcutaneous preadipocytes
The successfully constructed pcDNA-3.1-STEAP4 plasmid (Fig. 6a) was transferred into goat subcutaneous pre-adipocytes. STEAP4 expression was detected to be up-regulated by about 8900 times (Fig. 6b), indicating the successful establishment of STEAP4 cell overexpression model. Using BODIPY and Oil Red O staining methods to observe the formation of intracellular lipid droplets, it was found that overexpression of STEAP4 promoted lipid droplet accumulation in subcutaneous adipocytes (Fig. 6c, d, e). Detection of the expression of differentiation marker genes showed that C/EBPβ (CCAAT enhancer binding protein) was extremely significantly up-regulated (P<0.01), aP2 was significantly up-regulated (P<0.05), PPARγ (peroxisome proliferator-activated receptor) and Pref-1 (the preadipocyte factor) were significantly down-regulated (P<0.05), and LPL (lipoprotein lipase) was extremely significantly down-regulated (P<0.01).
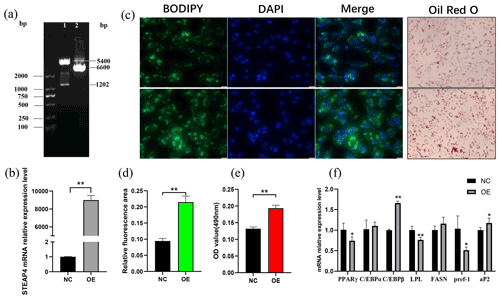
Figure 6The effect of overexpression of goat STEAP4 on the differentiation of subcutaneous preadipocytes. (a) pcDNA-3.1-STEAP4 restriction digestion diagram. (b) Efficiency of STEAP4 overexpression. (c) Image of BODIPY and Oil Red O staining. (d) BODIPY fluorescence quantification. (e) Oil Red O staining lipid droplet quantification. (f) Adipocytes differentiation-related gene expression. “” means P<0.01, and “*” means P<0.05.
STEAPs can play an important role in growth and development and the occurrence of various diseases by regulating their target genes (Qin et al., 2010). In this study, we cloned the goat STEAP4 sequence of 1388 bp, including the complete CDS of 1197 bp. The goat STEAP4 gene collectively encoded 398 amino acids, while human STEAP4 contains 459 amino acids, sheep STEAP4 contains 384 amino acids, cattle contains 470 amino acids, and pig STEAP4 also contains 470 amino acids (Wang et al., 2013), indicating that the length and composition of the amino acid sequence of STEAP4 differ among different species. This is compared with the predicted sequence in NCBI (XM_005679300.3). There were three base mutations in the CDS region of the goat STEAP4 gene, which are A188G, T281C, and A507G. A507G changed the amino acid at position 170 from Ile to Val. Whether it is unique to the Jianzhou Da'er goat and whether the mutation causes changes in protein function still needs further research.
This study used online tools to predict that goat STEAP4 was an stable hydrophilic basic protein with a molecular mass of 45.327 kD and a theoretical isoelectric point (pI) of 9.08. The subcellular location showed that STEAP4 was mainly located in the plasma membrane, which was consistent with a number of studies (Zhang et al., 2008; Wellen et al., 2007; Xue et al., 2017) and conforms to the membrane location characteristics of the STEAP family. Different tools were used to predict that goat STEAP4 contains four- transmembrane domains, which was different from the six-transmembrane structure conserved in the STEAP family. The predecessor obtained a variant of STEAP4, STEAP4v, when cloning the pig STEAP4 gene, which contained only four-transmembrane structure regions (Wang et al., 2013). The results of this study were similar to it. Therefore, we speculated that there may be cleavage of goat STEAP4 at the post-translational level, which leads to truncation of goat STEAP4. But what kind of functional changes may cause this cleavage still needs further study.
The goat STEAP4 gene was expressed in pancreas, heart, spleen, kidney, back subcutaneous fat tissue, abdominal subcutaneous fat tissue, liver, lung, and longissimus doris. But the relative expression level was the highest in the liver, followed by the lung. There were also higher expressions in the subcutaneous fat tissues of different parts. Moldes et al. (2001) reported that STEAP4 was expressed in large amounts in rat white fat, brown fat, heart, liver, and muscle. Human STEAP4 was highly expressed in bone marrow, placenta, fetal liver, and adipose tissue (Korkmaz et al., 2005). Pig STEAP4 was highly expressed in adipose tissue, kidney, cerebellum, heart, and lung, while STEAP4v was highly expressed in lung and adipose tissue (Wang et al., 2013). Combined, the results of this experiment show that STEAP4 was expressed at a high level in adipose tissues of different species, indicating that the tissue distribution of STEAP4 had a certain consistency, and there were also species differences.
The high expression of STEAP4 in adipose tissue suggested that it may regulate lipid accumulation in animals. Therefore, this study continues to detect the expression of STEAP4 in goat subcutaneous adipocytes. The results showed that STEAP4 was widely expressed during adipocyte differentiation and showed a decreasing and then an increasing trend, which decreased in the early stage of differentiation, and then it showed a trend of continuous increase. In 3T3-L1 preadipocytes, STEAP4 showed an upward trend with differentiation (Moldes et al., 2001), and the results of this experiment were similar. The expression trend in human adipocytes was opposite to that of 3T3-L1 and goat subcutaneous adipocytes. Human STEAP4 was highly expressed in preadipocytes, and its expression decreased with the induction of differentiation, being its lowest when adipocytes were mature (Chen et al., 2010). This may be due to differences in species and cell types. The above results suggested that STEAP4 may have different roles in the differentiation process of adipocytes of different species, and the role of STEAP4 in the differentiation process of goat subcutaneous adipocytes still needs further study.
Therefore, this study constructed a goat STEAP4 expression vector. After overexpression, it was observed that the accumulation of intracellular lipid droplets increased, and the mRNA expression of adipogenic differentiation marker genes, CCAAT enhancer binding protein (C/EBPβ), and fatty acid binding protein (aP2) was extremely and significantly up-regulated. C/EBPα and fatty acid synthase (FASN) mRNA levels also showed an upward trend. The expression of the preadipocyte factor (Pref-1), the peroxisome proliferator-activated receptor (PPARγ), and lipoprotein lipase (LPL) was down-regulated. Adipocyte differentiation was a complex process involving the coordinated action of multiple transcription factors. A number of studies have shown that C/EBPs and PPARs played a central role in adipocyte differentiation (Morrison and Farmer, 1999; Rosen and Spiegelman, 2000; Shao and Lazar, 1997). There was also a cascade of expression between them. C/EBPβ was necessary for the early stage of adipogenesis, and its expression could activate PPARγ and C/EBPα (Schwarz et al., 1997; Park et al., 2004; Ayala-Sumuano et al., 2008). In this experiment, C/EBPβ was extremely significantly up-regulated after STEAP4 overexpression, and C/EBPα also showed an up-regulation trend, suggesting that the cells were in the process of transitioning from the initial stage of differentiation. Pref-1 was a marker of preadipocytes, which could inhibit adipocyte differentiation to maintain the phenotype of preadipocytes (Smas and Sul, 1993). LPL regulated systemic lipid distribution and can hydrolyze triglycerides (Merkel and Eckel, 2002). FASN participated in fatty acid synthesis (Song et al., 2018), and aP2 was a fatty acid binding protein and the main target gene of PPARγ (Boord et al., 2002; Furuhashi and Hotamisligil, 2008). In addition, aP2 can also negatively regulate the level of PPARγ (Garin-Shkolnik, 2014). So, the down-regulation of PPARγ in this experiment may be negatively regulated by the up-regulation of aP2. Combined with the experimental results of this study, it was speculated that the fatty acid synthesis increases, and the hydrolysis pathway was blocked after the overexpression of STEAP4, which leads to the accumulation of lipid droplets. According to reports by Korkmaz et al. (2005), C/EBPα can directly bind to and promote the promoter activity of human and mouse STEAP4, while PPARγ does not directly act on the promoter of STEAP4. But whether goat STEAP4 was directly regulated by C/EBPs or PPARs to promote the differentiation of goat subcutaneous adipocytes still needs further study.
In this study, the goat STEAP4 gene sequence containing the complete CDS region was cloned. The protein physicochemical analysis showed that it was a basic hydrophilic protein. Goat STEAP4 has the highest amino acid sequence identity with sheep and has the closest genetic relationship with sheep. After overexpression of STEAP4 in goat, it was comprehensively demonstrated through morphology and molecular biology that overexpression positively regulated the differentiation of preadipocytes.
The original data are available upon request to the corresponding author.
YaqL designed and supervised the study. XL, HZ, JZ, and YW curated and analyzed the data. YanL and YX managed the figures. XL wrote the paper. YaqL and RL reviewed and revised the paper. All authors have read and agreed to the published version of the paper.
The contact author has declared that none of the authors has any competing interests.
All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee, Southwest Minzu University (Chengdu, Sichuan, China).
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors thank Wenlin Bai for his assistance in the revision of this article.
This research has been supported by the National Natural Science Foundation of China (grant nos. 32072723, 31672395, and 82002817), the Applied Basic Research Program of Sichuan Province (grant no. 2021YFYZ0003), and the Fundamental Research Funds for the Central Universities (grant no. 2021057).
This paper was edited by Henry Reyer and reviewed by Wenlin Bai and two anonymous referees.
Ayala-Sumuano, J. T., Velez-Del, Valle, C., Beltrán-Langarica, A., Hernández, J. M., and Kuri-Harcuch, W.: Adipogenic genes on induction and stabilization of commitment to adipose conversion, Biochem. Bioph. Res. Co., 374, 720–724, https://doi.org/10.1016/j.bbrc.2008.07.127, 2008.
Boord, J. B., Fazio, S., and Linton, M. F.: Cytoplasmic fatty acid-binding proteins: emerging roles in metabolism and atherosclerosis, Curr. Opin. Lipidol., 13, 141–147, https://doi.org/10.1097/00041433-200204000-00005, 2002.
Chen, X. H., Zhu, C., Ji, C. B., Zhao, Y. P., Zhang, C. M., Chen, F. K., Gao, C. L., Zhu, J. G., Qian, L. M., and Guo, X. R.: STEAP4, a gene associated with insulin sensitivity, is regulated by several adipokines in human adipocytes, Int. J. Mol. Med., 25, 361–367, https://doi.org/10.3892/ijmm_00000353, 2010.
Furuhashi, M. and Hotamisligil, G. S.: Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets, Nat. Rev. Drug Discov., 7, 489–503, https://doi.org/10.1038/nrd2589, 2008.
Garin-Shkolnik, T., Rudich, A., Hotamisligil, G. S., and Rubinstein, M.: FABP4 attenuates PPARγ and adipogenesis and is inversely correlated with PPARγ in adipose tissues, Diabetes, 63, 900–911, https://doi.org/10.2337/db13-0436, 2014.
He, C. S., Wang, Y., Xu, Q., Xiong, Y., Zhu, J. J., and Lin, Y. Q.: Overexpression of Kruppel like factor 3 promotes subcutaneous adipocytes differentiation in goat (Capra hircus), Anim. Sci. J., 92, e13514, https://doi.org/10.1111/asj.13514, 2021.
Hubert, R. S., Vivanco, I., Chen, E., Rastegar, S., Leong, K., Mitchell, S. C., Madraswala, R., Zhou, Y., Kuo, J., Raitano, A. B., Jakobovits, A., Saffran, D. C., and Afar, D. E.: STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors, P. Natl. Acad. Sci. USA, 96, 14523–14528, https://doi.org/10.1073/pnas.96.25.14523, 1999.
Kim, K., Mitra, S., Wu, G., Berka, V., Song, J., Yu, Y., Poget, S., Wang, D. N., Tsai, A. L., and Zhou, M.: Six-Transmembrane Epithelial Antigen of Prostate 1 (STEAP1) Has a Single b Heme and Is Capable of Reducing Metal Ion Complexes and Oxygen, Biochemistry, 55, 6673–6684, https://doi.org/10.1021/acs.biochem.6b00610, 2016.
Kong, Y. Y., Zhang, X. Y., Li, F. D., and Yue, X. P.: Research Progress on the Relationship Between Mutton Flavor Precursor Substance and Mutton Flavor and Influencing Factors, Journal of Agricultural Biotechnology, 29, 1612–1621, 2021.
Korkmaz, C. G., Korkmaz, K. S., Kurys, P., Elbi, C., Wang, L., Klokk, T. I., Hammarstrom, C., Troen, G., Svindland, A., Hager, G. L., and Saatcioglu, F.: Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer, Oncogene, 24, 4934–4945, https://doi.org/10.1038/sj.onc.1208677, 2005.
Lefterova, M. I. and Lazar, M. A.: New developments in adipogenesis, Trends Endocrin. Met., 20, 107–114, https://doi.org/10.1016/j.tem.2008.11.005, 2009.
Merkel, M. and Eckel, R. H., and Goldberg, I. J.: Lipoprotein lipase: genetics, lipid uptake, and regulation, J. Lipid Res., 43, 1997–2006, https://doi.org/10.1194/jlr.r200015-jlr200, 2002.
Millon-Frémillon, A., Bouvard, D., Grichine, A., Manet-Dupé, S., Block, M. R., and Albiges-Rizo, C.: Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity, J. Cell Biol., 180, 427–441, https://doi.org/10.1083/jcb.200707142, 2008.
Moldes, M., Lasnier, F., Gauthereau, X., Klein, C., Pairault, J., Fève, B., and Chambaut-Guérin, A. M.: Tumor necrosis factor-alpha-induced adipose-related protein (TIARP), a cell-surface protein that is highly induced by tumor necrosis factor-alpha and adipose conversion, J. Biol. Chem., 276, 33938–33946, https://doi.org/10.1074/jbc.M105726200, 2001.
Morrison, R. F. and Farmer, S. R.: Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis, J. Biol. Chem., 274, 17088–17097, https://doi.org/10.1074/jbc.274.24.17088, 1999.
Ohgami, R. S., Campagna, D. R., Greer, E. L., Antiochos, B., McDonald, A., Chen, J., Sharp, J. J., Fujiwara, Y., Barker, J. E., and Fleming, M. D.: Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells, Nat. Genet., 37, 1264–1269, https://doi.org/10.1038/ng1658, 2005.
Ohgami, R. S., Campagna, D. R., McDonald, A., and Fleming, M. D.: The Steap proteins are metalloreductases, Blood, 108, 1388–1394, https://doi.org/10.1182/blood-2006-02-003681, 2006.
Oosterheert, W., van Bezouwen, L. S., Rodenburg, R. N. P., Granneman, J., Förster, F., Mattevi, A., and Gros, P.: Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction, Nat. Commun., 9, 4337, https://doi.org/10.1038/s41467-018-06817-7, 2018.
Ozmen, F., Ozmen, M. M., Gelecek, S., Bilgic, İ., Moran, M., and Sahin, T. T.: STEAP4 and HIF-1α gene expressions in visceral and subcutaneous adipose tissue of the morbidly obese patients, Mol. Immunol., 73, 53–59, https://doi.org/10.1016/j.molimm.2016.03.008, 2016.
Park, B. H., Qiang, L., and Farmer, S. R.: Phosphorylation of C/EBPbeta at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes, Mol. Cell Biol., 24, 8671–8680, https://doi.org/10.1128/MCB.24.19.8671-8680.2004, 2004.
Qin, D. N., Kou, C. Z., Ni, Y. H., Zhang, C. M., Zhu, J. G., Zhu, C., Wang, Y. P., Zhu, G. Z., Shi, C., Ji, C. B., and Guo, X. R.: Monoclonal antibody to the six-transmembrane epithelial antigen of prostate 4 promotes apoptosis and inhibits proliferation and glucose uptake in human adipocytes, Int. J. Mol. Med., 26, 803–811, https://doi.org/10.3892/ijmm_00000528, 2010.
Rosen, E. D. and Spiegelman, B. M.: Molecular regulation of adipogenesis, Annu. Rev. Cell Dev. Biol., 16, 145–171, https://doi.org/10.1146/annurev.cellbio.16.1.145, 2000.
Schwarz, E. J., Reginato, M. J., Shao, D., Krakow, S. L., and Lazar, M. A.: Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription, Mol. Cell Biol., 17, 1552–1561, https://doi.org/10.1128/MCB.17.3.1552, 1997.
Shao, D. and Lazar, M. A.: Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation, J. Biol. Chem., 272, 21473–21478, https://doi.org/10.1074/jbc.272.34.21473, 1997.
Smas, C. M. and Sul, H. S.: Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation, Cell, 73, 725–734, https://doi.org/10.1016/0092-8674(93)90252-l, 1993.
Song, Z., Xiaoli, A. M., and Yang, F.: Regulation and Metabolic Significance of De Novo Lipogenesis in Adipose Tissues, Nutrients, 10, 1383, https://doi.org/10.3390/nu10101383, 2018.
Tanaka, Y., Matsumoto, I., Iwanami, K., Inoue, A., Minami, R., Umeda, N., Kanamori, A., Ochiai, N., Miyazawa, K., Sugihara, M., Hayashi, T., Goto, D., Ito, S., and Sumida, T.: Six-transmembrane epithelial antigen of prostate4 (STEAP4) is a tumor necrosis factor alpha-induced protein that regulates IL-6, IL-8, and cell proliferation in synovium from patients with rheumatoid arthritis, Mod. Rheumatol., 22, 128–136, https://doi.org/10.1007/s10165-011-0475-y, 2012.
Valenti, M. T., Giannini, S., Donatelli, L., Realdi, G., Lo Cascio, V., and Dalle Carbonare, L.: Zoledronic acid decreases mRNA six-transmembrane epithelial antigen of prostate protein expression in prostate cancer cells, J. Endocrinol. Invest., 33, 244–249, https://doi.org/10.1007/BF03345787, 2010.
Wang, S. B., Lei, T., Zhou, L. L., Zheng, H. L., Zeng, C. P., Liu, N., Yang, Z. Q., and Chen, X. D.: Functional analysis and transcriptional regulation of porcine six transmembrane epithelial antigen of prostate 4 (STEAP4) gene and its novel variant in hepatocytes, Int. J. Biochem. Cell B., 45, 612–620, https://doi.org/10.1016/j.biocel.2012.12.011, 2013.
Wellen, K. E., Fucho, R., Gregor, M. F., Furuhashi, M., Morgan, C., Lindstad, T., Vaillancourt, E., Gorgun, C. Z., Saatcioglu, F., and Hotamisligil, G. S.: Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis, Cell, 129, 537–548, https://doi.org/10.1016/j.cell.2007.02.049, 2007.
Xue, X., Bredell, B. X., Anderson, E. R., Martin, A., Mays, C., Nagao-Kitamoto, H., Huang, S., Győrffy, B., Greenson, J. K., Hardiman, K., Spence, J. R., Kamada, N., and Shah, Y. M.: Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer, P. Natl. Acad. Sci. USA, 114, E9608–E9617, https://doi.org/10.1073/pnas.1712946114, 2017.
Yan, D., Dong, W., He, Q., Yang, M., Huang, L., Kong, J., Qin, H., Lin, T., and Huang, J.: Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation, EBioMedicine, 48, 316–331, https://doi.org/10.1016/j.ebiom.2019.08.074, 2019.
Zhang, C. M., Chi, X., Wang, B., Zhang, M., Ni, Y. H., Chen, R. H., Li, X. N., and Guo, X. R.: Downregulation of STEAP4, a highly-expressed TNF-alpha-inducible gene in adipose tissue, is associated with obesity in humans, Acta Pharm. Sinic., 29, 587–592, https://doi.org/10.1111/j.1745-7254.2008.00793.x, 2008.