the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Expression analysis and single-nucleotide polymorphisms of SYNDIG1L and UNC13C genes associated with thoracic vertebral numbers in sheep (Ovis aries)
Ying-Jie Zhong
Yang Yang
Xiang-Yu Wang
Ran Di
Ming-Xing Chu
Qiu-Yue Liu
The objective of the current study was to analyze expression levels of synapse differentiation inducing 1-like (SYNDIG1L) and unc-13 homolog C (UNC13C) genes in different tissues, while single-nucleotide polymorphisms (SNPs) of two genes were associated with multiple thoracic vertebrae traits in both Small-tailed Han sheep (STH) and Sunite sheep (SNT). The expression levels of SYNDIG1L and UNC13C were analyzed in the brain, cerebellum, heart, liver, spleen, lung, kidney, adrenal gland, uterine horn, longissimus muscle, and abdominal adipose tissues of two sheep breeds with different thoracic vertebral number (TVN) sheep (T13 groups and T14 groups) by real-time quantitative polymerase chain reaction (RT-qPCR). Meanwhile, the polymorphisms of UNC13C gene g.52919279C>T and SYNDIG1L gene g.82573325C>A in T14 and T13 were genotyped by the Sequenom MassARRAY® SNP assay, and association analysis was performed with the TVN. The results demonstrated that UNC13C gene was extensively expressed in 11 tissues. The expression of UNC13C gene in longissimus muscle of T14 groups of STH was significantly higher than that of T13 groups (P<0.05). SYNDIG1L gene was overexpressed in brain and cerebellum tissues, and the expression level of UNC13C gene in the brain and cerebellum of T13 groups in SNT was significantly higher than that of T14 groups (P<0.01). Association analysis showed that SNPs found in the UNC13C gene had no significant effects on TVN for both two genes. The polymorphism of SYNDIG1L g.82573325C>A was significantly correlated with the TVN in both STH (P<0.05) and SNT (P<0.01). Taken together, the SYNDIG1L gene was related to thoracic vertebral development, and this variation may be potentially used as a molecular marker to select the multiple thoracic vertebrae in sheep.
- Article
(814 KB) - Full-text XML
- BibTeX
- EndNote
The spine of a vertebrate consists of a series of repeated vertebrae. Based on morphological differences, the vertebrae were subdivided into five distinct functional spinal regions: cervical, thoracic, lumbar, sacral, and caudal (Donaldson et al., 2013). The number of vertebrae is relatively conserved among mammalian species. However, the quantitative variations of vertebrae (Sun et al., 2019) have been observed in pigs (Rohrer and Nonneman, 2017), deer (Mizer and Wahl, 2018), humans (Ibrahim et al., 2013), and sheep (Donaldson et al., 2013). In general, the vertebrae of sheep were arranged from the neck to the sacral part according to 7 cervical vertebrae (C), 13 thoracic vertebrae (T), 6 lumbar vertebrae (L), and 4 sacral vertebrae (S), with a total of 30 vertebrae. Among them, mutations in the thoracolumbar position were the most common (T14L6 or T13L7) (Zhang, 1996). Multi-vertebrae sheep have advantages in adaptability and meat production performance (Zhang et al., 1996). The cultivation of multi-spine sheep has comprehensive benefits for the economy, society, and ecology. This is of great significance to improve the quality and efficiency of the animal husbandry industry.
Among domestic animals, the most extensive studies of vertebral number variation have focused on pigs. Previous studies have reported quantitative trait loci (QTL) for vertebral numbers in pigs by genome scans based on microsatellite markers. Two genome-wide significant QTLs were detected on pig chromosomes (SSCs) 1 and 2 in a Meishan and Göttingen cross line (Wada et al., 2000). The vertebrae-development-associated (VRTN) gene on the SSC7 and NR6A1 gene on SSC1 were considered as candidate genes affecting vertebral numbers. Fine mapping of vertebral number trait was performed, and an orphan nuclear receptor, germ cell nuclear factor (NR6A1) was localized to be the candidate and also confirmed by multiple studies (Mikawa et al., 2011; Zhang et al., 2015), which was confirmed in various studies (Fan et al., 2013; Rohrer et al., 2015; Yang et al., 2016). However, current studies on sheep vertebral number traits were superficial, and functional studies focused on genomic variations were relatively rare (Cao et al., 2015; Chen et al., 2012).
Small-tailed Han sheep (STH) selected in this study is one of the famous indigenous sheep breeds of China which grows fast and has good early puberty and high fecundity (Guo et al., 2020). Sunite sheep (SNT) are also an indigenous breed that has the advantages of cold/drought resistance, fast growth, and good disease resistance. Meat production performance is good, lean meat percentage is high, protein content is high, it has a high popularity, and it has a large demand space at home and even abroad (Zhong et al., 2020; Gao et al., 2014). There was a high proportion of multiple thoracic and lumbar vertebral numbers in both sheep breeds. Identification of molecular markers of multi-spine variation for marker-assisted selection is of great significance for the improvement of meat production performance.
Previously, we conducted a genome-wide association analysis in two sheep breeds of 670 sheep with different thoracic vertebra numbers using a Affymetrix ovine 600K single-nucleotide polymorphism (SNP) array. Genome-wide significant associations were detected at nine SNPs in the 245 kb region with (Yingjie Zhong, unpublished data). The significant SNPs on chromosome 7 were located near the region of synapse differentiation inducing 1-like (SYNDIG1L) and unc-13 homolog C (UNC13C). Whole-genome resequencing was also performed on 40 sheep with thoracic vertebra numbers for fine mapping. Using the top 10 % of Fst values as cutoffs, candidate genes associated with thoracic vertebra number were identified, which included SYNDIG1L and UNC13C genes. Annotation of the sheep reference genome (Oar4.0) suggested that the two non-synonymous mutations are located in protein-coding regions of synapse differentiation inducing 1-like SYNDIG1L and UNC13C genes respectively. g.82573325C>A located on exon 3 of SYNDIG1L is a non-synonymous mutation that changes amino acid position 186 from glycine (G) to tryptophan (W), while UNC13C g.52919279C>T located on exon 14 is also a non-synonymous mutation that changes amino acid position 1465 from valine (V) to isoleucine (I).
The purpose of this study is to explore the association of UNC13C g.52919279C>T and SYNDIG1L g.82573325C>A loci with thoracic vertebral number. It provides promising candidate causal mutations for further research on the number of vertebral variations on sheep.
2.1 Animal and main reagent
All the experimental procedures mentioned in the present study were approved by the Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS) (Beijing, China). In addition, there was ethics approval by the animal ethics committee of IAS-CAAS (no. IAS2020-82, 28 July 2020).
A total of 12 healthy ewes aged 3 years old were selected from the livestock and breeding base of Tianjin Animal Husbandry and Veterinary Research Institute. The number of thoracic vertebrae of SNT and STH was 13 and 14, respectively. After slaughter, 11 tissues of brain, cerebellum, heart, liver, spleen, lung, kidney, adrenal gland, uterine horn, longissimus muscle, and abdominal fat were quickly collected, put into a 2 mL RNase-free centrifuge tube, and stored in liquid nitrogen immediately. After returning to the laboratory, they were stored in the freezer at −80 ∘C.
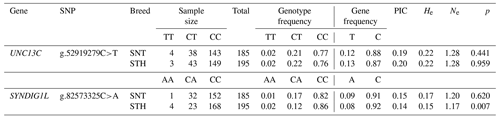
For genotyping (Table 1), a total of 383 sheep were selected from SNT and STH from Bayan Nur slaughterhouses in the Inner Mongolia autonomous region, China, and Yuncheng slaughterhouses in Shandong province. After slaughter, the collected fresh muscle tissue was quickly put into a 2 mL frozen storage tube and immediately stored in liquid nitrogen. After being brought back to the laboratory, all the fresh muscle tissue was transferred to a freezer at −80 ∘C for storage.
2.2 Extraction of genomic DNA and total RNA and main reagents
DNA from muscle tissue was extracted by a DNA extraction kit (TIANGEN Biotech, Beijing, China). The total RNA of tissue was extracted using the Trizol and Qiagen RNeasy kit (Qiagen). The concentration and integrity of DNA and RNA were detected by Nanodrop2000, and the quality of DNA and RNA was detected by 1.5 % agarose gel electrophoresis.
Quantitative polymerase chain reaction (PCR) was done using the SYBR Green fluorescent dye for product detection (SYBR® Premix Ex Taq™ II). The PrimeScript™ RT reagent kit was used to synthesize cDNA (TaKaRa, Beijing).
2.3 cDNA synthesis
The total volume of the reaction system was 20 µL / 4.0 µL 5× PrimeScript buffer (for real time), 1.0 µL PrimeScript RT enzyme mix E, 1.0 µL Oligo dT primer, 1.0 µL random 6 mers, 1000 ng RNA. The remaining system was supplemented with RNase-free ddH2O. The reaction condition of PCR was 37 ∘C for 15 min and 85 ∘C for 5 s. The product obtained after reverse transcription was diluted five times and stored in a freezer at −20 ∘C for detection of tissue expression of the target gene.
2.4 Primer design
Primers for real-time quantitative polymerase chain reaction (RT-qPCR) were designed using the GenBank database (https://www.ncbi.nlm.nih.gov/genbank, last access: 10 July 2020). Genes and their accession numbers include SYNDIG1L (GenBank: XM_027972017.1), UNC13C (GenBank: XM_027971817.1). The β-actin (GenBank: NM_001009784.2) was an internal reference gene. The primers (Table 2) were synthesized by Beijing Tianyi Huiyuan Biotechnology Co., Ltd.
2.5 Real-time fluorescent quantitative PCR
Using a Roche Light Cycler® 480 type II fluorescence quantitative PCR instrument, the whole PCR process was monitored in real time by fluorescence signal accumulation, and β-actin was used as an internal reference gene. The total volume of the reaction system was 20 µL: SYBR Premix Ex Taq II 10 µL, forward primer 0.8 µL, reverse primer 0.8 µL, RNase-free ddH2O 6.4 µL, and cDNA 2.0 µL. PCR conditions were as follows: initial denaturation at 95 ∘C for 5 s, followed by 40 cycles of 95 ∘C for 10 s and 60 ∘C for 30 s.
2.6 Genotyping
Genotyping of UNC13C g.52919279C>T and SYNDIG1L g.82573325C>A was carried out using the Sequenom MassARRAY® SNP (Johansen et al., 2013; Ortega et al., 2017) assay. The primer information is provided in Table 3. The typing sample is DNA, and the amount required for each sample is 20 µL. DNA concentration ranged from 40 to 80 ng/ µL.
2.7 Statistical analysis
The relative expressions of UNC13C and SYNDIG1L were calculated by the 2−ΔΔCt method. The difference of relative expression between the T13 group and the T14 group was analyzed by one-way ANOVA. The allele frequency, genotype frequency, p value, polymorphism information content (PIC), heterozygosity (He), and effective allele number (Ne) were calculated using Microsoft Excel 2016 statistical software. Then, the distribution of genotypes for each SNP in the studied populations was tested for deviation from Hardy–Weinberg equilibrium. P>0.05 indicates the locus was under Hardy–Weinberg equilibrium.
The correlation between SNP and thoracic vertebra number traits of two varieties was analyzed by SAS (V.9.4) (SAS Institute Inc.). The p values < 0.05 were considered to be significant. The mathematical models are as follows: Fisher's exact probability test and logistic regression.
Here, y represents the number of thoracic vertebrae, X1 represents the variety, and X2 represents the genotype.
3.1 Polymorphism analysis of SYNDIG1L and UNC13C genes
The genotyping results of 383 sheep (Fig. 1) showed that two candidate loci were polymorphic (Table 4). UNC13C g.52919279C>T and SYNDIG1L g.82573325C>A displayed low polymorphisms (PIC < 0.25) in both SNT and STH populations. Statistical significance was analyzed by the chi-square test. The frequency of SYNDIG1L g.82573325C>A was consistent with Hardy–Weinberg equilibrium in SNT, ensuring the reliability of their application to evaluating larger groups (P>0.05). SYNDIG1L g.82573325C>A was, however, under Hardy–Weinberg imbalance (P<0.05) in the STH population. UNC13C g.52919279C>T satisfied the Hardy–Weinberg equilibrium in both populations (P>0.05).
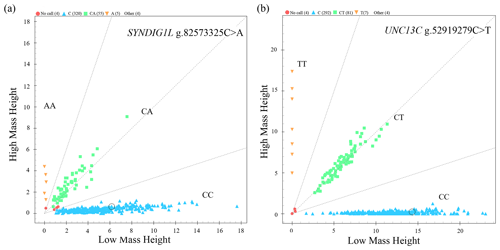
Figure 1Genotyping results. (a) Scatter plot of SYNDIG1L genotyping results. (b) Scatter plot of UNC3C genotyping results.
3.2 Association analysis of SYNDIG1Land UNC13C genes with thoracic vertebral number in two breeds
Firstly, association analysis of SNPs with thoracic vertebral number (TVN) was explored in two sheep breeds, respectively. The statistical results were shown in Table 5. UNC13C g.52919279C>T had no significant effect on TVN in both the STH and SNT populations (P>0.05). SYNDIG1L g.82573325C>A was significantly correlated with different thoracic vertebral numbers in both STH (P<0.05) and SNT (P<0.01). Then, logistic regression was used to test the effects of breeds and genotypes on different thoracic vertebral numbers in sheep, which was shown in Table 6. For the two candidate SNPs, the breeds have no significant relevance with TVN in sheep (P>0.05). The genotypes of SYNDIG1L g.82573325C>A were significantly associated with multiple thoracic vertebrae in sheep (P<0.01), indicating that this gene might be correlated with a TVN trait in the sheep.
Table 5Genotypes of candidate locus and the number of thoracic vertebrae in a single breed by Fisher's exact test.
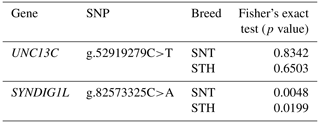
P≤0.05 indicates the significant difference; P≤0.01 indicates the extremely significant difference.
3.3 Expression profiles of UNC13C and SYNDIG1L genes in SNT and STH with different TVNs
The results of RT-qPCR showed that the UNC13C gene was extensively expressed in 11 tissues of STH and SNT (Fig. 2a). The SYNDIG1L gene was mainly expressed in brain tissue and slightly expressed in the spleen in SNT with T13 (Fig. 2b). The UNC13C gene was highly expressed in the cerebellum of STH. The expression of UNC13C in group T14STH was significantly higher than group T13STH in the longissimus muscle (P<0.05); the gene expression fold change is 1.8. In T13SNT, the expression of the SYNDIG1L gene in the brain and cerebellum tissues was significantly higher than that in T14SNT (P<0.05); the fold changes of gene expression were 0.5 and 0.4, respectively.
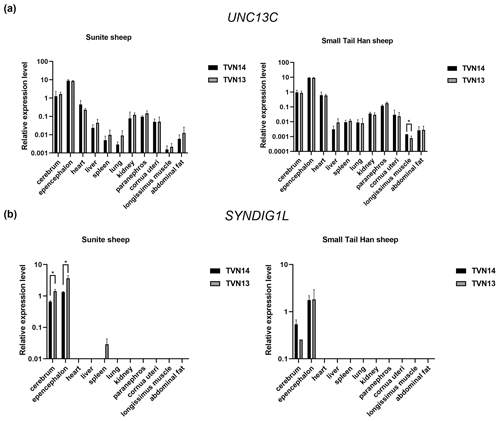
Figure 2Results of expression level of UNC13C and SYNDIG1L genes in STH and SNT with different TVNs. (a) The comparison of expression levels of UNC3C between SNT and STH. (b) The comparison of expression levels of SYNDIG1L between SNT and STH. The significant results with a p value lower than 0.05 are given one asterisk (∗).
The vertebrae of mammals are derived from the mesoderm of the gastrula. Vertebrae development is an extremely complicated system that is regulated temporally and spatially. It has been known that any error in development can result in many congenital abnormalities (Gilbert, 2003). Zhang et al. (1998) found that the meat-production performance of multi-vertebrae sheep was significantly better than that of normal sheep, with longer longissimus muscle, larger abdominal cavity volume, carcass weight, net meat weight, lean meat percentage, and other economic indexes. Moreover, this trait is heritable. Thus, it is important to understand the mechanism of vertebral number variation from the molecular level and apply it to sheep breeding with multiple thoracic vertebrae. The genetic architecture of thoracic vertebral number has been extensively studied in pigs, and major genes affecting this trait have been mapped in indigenous pigs (Rohrer et al., 2015; Duan et al., 2018; Liu et al., 2020). However, current studies on sheep vertebral numbers were superficial, and functional studies focused on genomic variations were relatively rare.
Liu et al. (2020) found that regulation variants on SSC7 might play crucial roles in the number of thoracic vertebrae (NTV) and the FOS (Fos proto-oncogene, AP-1 transcription factor subunit) on SSC7, and BMPR1A was identified as a novel candidate gene affecting the NTV in pigs on SSC14. Fan et al. (2013) identified three loci for a vertebral number trait through a genome-wide association study and located them in a 947 kb region on SSC7 in pigs. The locus was refined to 100 kb by a homologous sharing test, which contained only VRTN and SYNDIG1L genes. Among them, VRTN is considered to be the main candidate gene affecting vertebral numbers in the modern western world. The VRTN gene was considered as a candidate gene affecting vertebral number also in sheep (Li et al., 2019). We believe that it is not accidental that SYNDIG1L and VRTN have been identified at the same time. SYNDIG1L is highly expressed in the striatum (de Chaldée et al., 2006). This is consistent with our previous RT-qPCR results. As one of the basal ganglia of the brain, the striatum is mainly responsible for regulating muscle tension and coordinating various fine and complicated movements (Lorenc-Koci et al., 1998; Hemsley and Crocker, 2001). Meanwhile, it is related to the occurrence of Parkinson's disease (Miyanishi et al., 2019; Choe et al., 2011), chorea (Ishikawa et al., 1990), and other diseases. Correspondingly, some researchers found that the incidence of dyskinesia increased with the increase of thoracic vertebral numbers in pigs (Nakano et al., 2015). This may be due to the negative effects of high expression of SYNDIG1L. On the other hand, SYNDIG1L was reported to be a factor affecting the final body weight and back-fat thickness in Landrace pigs (Lee et al., 2018). An et al. (2020) believe that it is the key gene that affects the formation of bovine body shape. We speculate that SYNDIG1L may participate in the spine formation process and cause mutation in SYNDIG1L, which may lead to abnormal development of vertebrae in sheep. However, more evidence is needed to prove our hypothesis.
The new role of the UNC13C gene in oral squamous cell carcinoma (OSCC) has been revealed for the first time. UNC13C is a novel tumor suppressor and can be used as a target to prevent oral cancer metastasis (Velmurugan et al., 2019). Studies have shown that UNC13C is involved in Alzheimer's disease (AD), which involves dysfunction of many cellular pathways, including synaptic transmission, cytoskeleton dynamics, energetics, and apoptosis (Miller et al., 2013). According to references, UNC13C is significantly downregulated in spinal cord tissue of patients with amyotrophic lateral sclerosis (D'Erchia et al., 2017). It is considered to be negatively correlated with muscle ability in the study of myasthenia (Hangelbroek et al., 2016), no direct relationship between the function of the UNC13C gene and the number of thoracic vertebrae was found.
This study found that the polymorphisms of SYNDIG1L g.82573325C>A were significantly associated with the thoracic vertebral number in sheep, indicating that this locus may be a promising candidate causal variation in the regulation of thoracic vertebral numbers. Further exploration of the functions of the SYNDIG1L gene was necessary for the cultivation of sheep breeds with multiple thoracic vertebrae.
The data sets are available upon request from the corresponding authors.
YJZ and QYL contributed to the conception of the study. YY contributed significantly to analysis and manuscript preparation. YJZ performed the data analyses and wrote the manuscript. MXC contributed to revisions of the manuscript. XYW and RD assisted the analysis with constructive discussion.
The authors declare that they have no conflict of interest.
We thank Mingxing Chu, Qiuyue Liu, and all the faculties involved including Chinese Academy of Agricultural Sciences, as well as the local abattoir for their support during this study.
This work was supported by the Genetically Modified Organisms Breeding Major Program of China (grant no. 2016ZX08009-003-006), the Central Public-interest Scientific Institution Basal Research Fund (grant no. 2017ywf-zd-13), the Agricultural Science and Technology Innovation Program of China (grant no. ASTIP-IAS13), and the Earmarked Fund for China Agriculture Research System (grant no. CARS-38).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
An, B., Xu, L., Xia, J., Wang, X., Miao, J., Chang, T., Song, M., Ni, J., Xu, L., Zhang, L., Li, J., and Gao, H.: Multiple association analysis of loci and candidate genes that regulate body size at three growth stages in Simmental beef cattle, BMC Genet., 21, 32, https://doi.org/10.1186/s12863-020-0837-6, 2020.
Cao, J., Wei, C., Liu, D., Wang, H., Wu, M., Xie, Z., Capellini, T. D., Zhang, L., Zhao, F., Li, L., Zhong, T., Wang, L., Lu, J., Liu, R., Zhang, S., Du, Y., Zhang, H., and Du, L.: DNA methylation landscape of body size variation in sheep, Sci. Rep.-UK, 5, 13950, https://doi.org/10.1038/srep13950, 2015.
Chen, Q., Zhang, L. L., Zhao, J., and Ma, Y. H.: DNA methylation analysis of exon-1 of the ovine HOXC-8 gene in Mongolian sheep using bisulphite sequencing, J. Appl. Anim. Res., 40, 198–202, https://doi.org/10.1080/09712119.2012.658059, 2012.
Choe, M. A., An, G. J., Koo, B. S., and Jeon, S.: Effect of DHEA on recovery of muscle atrophy induced by Parkinson's disease, J. Korean Acad. Nurs., 41, 834–842, https://doi.org/10.4040/jkan.2011.41.6.834, 2011.
de Chaldée, M., Brochier, C., Van de Vel, A., Caudy, N., Luthi-Carter, R., Gaillard, M. C., and Elalouf, J. M.: Capucin: a novel striatal marker down-regulated in rodent models of Huntington disease, Genomics, 87, 200–207, https://doi.org/10.1016/j.ygeno.2005.10.009, 2006.
D'Erchia, A. M., Gallo, A., Manzari, C., Raho, S., Horner, D. S., Chiara, M., Valletti, A., Aiello, I., Mastropasqua, F., Ciaccia, L., Locatelli, F., Pisani, F., Nicchia, G. P., Svelto, M., Pesole, G., and Picardi, E.: Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS, Sci. Rep.-UK, 7, 10046, https://doi.org/10.1038/s41598-017-10488-7, 2017.
Donaldson, C. L., Lambe, N. R., Maltin, C. A., Knott, S., and Bunger, L.: Between- and within-breed variations of spine characteristics in sheep, J. Anim. Sci., 91, 995–1004, https://doi.org/10.2527/jas.2012-5456, 2013.
Duan, Y., Zhang, H., Zhang, Z., Gao, J., Yang, J., Wu, Z., Fan, Y., Xing, Y., Li, L., Xiao, S., Hou, Y., Ren, J., and Huang, L.: VRTN is required for the development of thoracic vertebrae in mammals, Int. J. Biol. Sci., 14, 667–681, https://doi.org/10.7150/ijbs.23815, 2018.
Fan, Y., Xing, Y., Zhang, Z., Ai, H., Ouyang, Z., Ouyang, J., Yang, M., Li, P., Chen, Y., Gao, J., Li, L., Huang, L., and Ren, J.: A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs, PLoS ONE, 8, e62534, https://doi.org/10.1371/journal.pone.0062534, 2013.
Gao, F. M., Bai, Y. E. T., Liu, J., Sun, Y. J., and Shao, Z. Y.: Quality and nutrition of Sunite sheep and mutton, Abstract of Animal Husbandry and Veterinary of China, 30, 44–43, https://doi.org/CNKI:SUN:ZXWA.0.2014-12-040, 2014.
Gilbert, S. F.: The morphogenesis of evolutionary developmental biology, Int. J. Dev. Biol., 47, 467–477, 2003.
Guo, D. E., Wang, W. K., Cui, B. H., Li, J., and Tang, l.: Comparison of production performance between Small tail Han sheep and Hu sheep, Jilin Animal Husbandry and Veterinary Medicine, 41, 60, 2020.
Hangelbroek, R. W., Fazelzadeh, P., Tieland, M., Boekschoten, M. V., Hooiveld, G. J., van Duynhoven, J. P., Timmons, J. A., Verdijk, L. B., de Groot, L. C., van Loon, L. J., and Müller, M.: Expression of protocadherin gamma in skeletal muscle tissue is associated with age and muscle weakness, J Cachexia Sarcopenia Muscle, 7, 604–614, https://doi.org/10.1002/jcsm.12099, 2016.
Hemsley, K. M. and Crocker, A. D.: Changes in muscle tone are regulated by D1 and D2 dopamine receptors in the ventral striatum and D1 receptors in the substantia nigra, Neuropsychopharmacology: Neuropsychopharmacology, 25, 514–526, https://doi.org/10.1016/s0893-133x(01)00245-7, 2001.
Ibrahim, D. A., Myung, K. S., and Skaggs, D. L.: Ten percent of patients with adolescent idiopathic scoliosis have variations in the number of thoracic or lumbar vertebrae, J. Bone Joint Surg. Am., 95, 828–833, https://doi.org/10.2106/jbjs.L.00461, 2013.
Ishikawa, A., Miyatani, N., Yuasa, T., Tanaka, K., and Oyanagi, K.: An autopsied case of manifesting chorea, serum antibody to brain proteins, neuronal degeneration in striatum and grumose degeneration in dentate nucleus, Rinsho Shinkeigaku, 30, 510–515, 1990.
Johansen, P., Andersen, J. D., Børsting, C., and Morling, N.: Evaluation of the iPLEX® Sample ID Plus Panel designed for the Sequenom MassARRAY® system. A SNP typing assay developed for human identification and sample tracking based on the SNPforID panel, Forensic science international, Forensic. Sci. Int. Genet., 7, 482–487, https://doi.org/10.1016/j.fsigen.2013.04.009, 2013.
Lee, Y. S., Shin, D., and Song, K. D.: Dominance effects of ion transport and ion transport regulator genes on the final weight and backfat thickness of Landrace pigs by dominance deviation analysis, Genes Genomics, 40, 1331–1338, https://doi.org/10.1007/s13258-018-0728-7, 2018.
Li, C. Y., Li, M., Li, X. Y., Ni, W., Xu, Y. R., Yao, R., Wei, B., Zhang, M. D., Li, H. X., Zhao, Y., Liu, L., Yaseen, U., Jiang, Y., and Hu, S. W.: Whole-genome resequencing reveals loci associated with thoracic vertebrae number in sheep, Front. Genet., 18, 674, https://doi.org/10.3389/fgene.2019.00674, 2019.
Liu, Q., Yue, J. W., Niu, N. Q., Liu, X., Yan, H., Zhao, F. P., Hou, X. H., Gao, H. M., Shi, L. J., Wang, L. X., Wang, L. G., and Zhang, L. C.: Genome-wide association analysis identified BMPR1A as a novel candidate gene affecting the number of thoracic vertebrae in a Large White × Minzhu intercross pig population. Animals (Basel), 10, 2186, https://doi.org/10.3390/ani10112186, 2020.
Lorenc-Koci, E., Konieczny, J., and Wolfarth, S.: Contribution of the glycine site of NMDA receptors in rostral and intermediate-caudal parts of the striatum to the regulation of muscle tone in rats, Brain Res., 793, 315–320, https://doi.org/10.1016/s0006-8993(98)00240-6, 1998.
Mikawa, S., Sato, S., Nii, M., Morozumi, T., Yoshioka, G., Imaeda, N., Yamaguchi, T., Hayashi, T., and Awata, T.: Identification of a second gene associated with variation in vertebral number in domestic pigs, BMC Genet., 12, 5, https://doi.org/10.1186/1471-2156-12-5, 2011.
Miller, J. A., Woltjer, R. L., Goodenbour, J. M., Horvath, S., and Geschwind, D. H.: Genes and pathways underlying regional and cell type changes in Alzheimer's disease, Genome Med., 5, 48, https://doi.org/10.1186/gm452, 2013.
Miyanishi, K., Choudhury, M. E., Watanabe, M., Kubo, M., Nomoto, M., Yano, H., and Tanaka, J.: Behavioral tests predicting striatal dopamine level in a rat hemi-Parkinson's disease model, Neurochem Int., 122, 38–46, https://doi.org/10.1016/j.neuint.2018.11.005, 2019.
Mizer, L. A. and Wahl, C.: The noncervical lateral transverse foramina, J Morphol, 279, 1679–1691, https://doi.org/10.1002/jmor.20905, 2018.
Nakano, H., Sato, S., Uemoto, Y., Kikuchi, T., Shibata, T., Kadowaki, H., Kobayashi, E., and Suzuki, K.: Effect of VRTN gene polymorphisms on Duroc pig production and carcass traits, and their genetic relationships, Anim. Sci. J., 86, 125–131, https://doi.org/10.1111/asj.12260, 2015.
Ortega, M. S., Denicol, A. C., Cole, J. B., Null, D. J., Taylor, J. F., Schnabel, R. D., and Hansen, P. J.: Association of single nucleotide polymorphisms in candidate genes previously related to genetic variation in fertility with phenotypic measurements of reproductive function in Holstein cows, J. Dairy Sci., 100, 3725–3734, https://doi.org/10.3168/jds.2016-12260, 2017.
Rohrer, G. A. and Nonneman, D. J.: Genetic analysis of teat number in pigs reveals some developmental pathways independent of vertebra number and several loci which only affect a specific side, Genet. Sel. Evol., 49, 4, https://doi.org/10.1186/s12711-016-0282-1, 2017.
Rohrer, G. A., Nonneman, D. J., Wiedmann, R. T., and Schneider, J. F.: A study of vertebra number in pigs confirms the association of vertnin and reveals additional QTL, BMC Genet., 16, 129, https://doi.org/10.1186/s12863-015-0286-9, 2015.
Sun, Q., Liu, S. J., Di, R., Hu, W. P., Wang, X. Y., Ma, L., Zhang, X. S., Zhang, J. L., Liu, Q. Y., and Chu, M. X.: Association between polymorphism of VRTN, NR6A1 genes and thoracic vertebral number variation and analysis of their tissue expression in Sunite sheep (Ovis aries), Journal of Agricultural Biotechnology, 27, 864–874, https://doi.org/CNKI:SUN:NYSB.0.2019-05-010, 2019.
Velmurugan, B. K., Yeh, K. T., Hsieh, M. J., Yeh, C. M., Lin, C. C., Kao, C. Y., Huang, L. R., and Lin, S. H.: UNC13C suppress tumor progression via inhibiting EMT pathway and improves survival in oral squamous cell carcinoma, Front Oncol., 9, 728, https://doi.org/10.3389/fonc.2019.00728, 2019.
Wada, Y., Akita, T., Awata, T., Furukawa, T., Sugai, N., Inage, Y., Ishii, K., Ito, Y., Kobayashi, E., Kusumoto, H., Matsumoto, T., Mikawa, S., Miyake, M., Murase, A., Shimanuki, S., Sugiyama, T., Uchida, Y., Yanai, S., and Yasue, H.: Quantitative trait loci (QTL) analysis in a Meishan x Göttingen cross population, Anim. Genet., 31, 376–384, https://doi.org/10.1046/j.1365-2052.2000.00696.x, 2000.
Yang, J., Huang, L., Yang, M., Fan, Y., Li, L., Fang, S., Deng, W., Cui, L., Zhang, Z., Ai, H., Wu, Z., Gao, J., and Ren, J.: Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs, Sci. Rep., 6, 19240, https://doi.org/10.1038/srep19240, 2016.
Zhang L. C., Liu X., Liang J., Yan H., Zhao K. B., LI N., PU L., Shi H. B., Zhang Y. B., Wang L. G.: Quantitative trait loci for the number of vertebrae on Sus scrofa chromosomes 1 and 7 independently influence the numbers of thoracic and lumbar vertebrae in pigs, J. Integr. Agr., 14, 2027–2033, https://doi.org/10.1016/S2095-3119(15)61084-X, 2015.
Zhang, L. L.: Genetic mechanism of polyspinous phenomena in Mongolian sheep, China Sheep, 6, 1–3, 1996.
Zhang, L. L., Ji, E. G. L., and Zhou, W.: Study on population Genetic characteristics of spinal number variation in Wuzhu Muqin sheep, Animal Husbandry and Feed Science, 000, 1–3, 1996.
Zhang, L. L., Luo, X. G., Si, Q. B. L. G., and Zhang, S. S.: Analysis of thoracolumar and lumbar spine length and meat production performance of Mongolian sheep with multiple vertebrae, Journal of Inner Mongolia Agricultural University (Natural Science Edition), 19, 1–5, 1998.
Zhong, Y. J., Xiang, G. M., He, X. Y., Liu, S. J., Zhang, X. S., Zhang, J. L., Chu, M. X., and Liu, Q. Y.: Preliminary study on the relationship between FBXL3 and FBXL21 gene expression and seasonal estrus in Sunite sheep, Chinese Journal of Animal Science, 56, 1–10, https://doi.org/10.19556/j.0258-7033.20200107-01, 2020.