the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Assessment of growth hormone gene polymorphism effects on reproductive traits in Holstein dairy cattle in Tunisia
Sihem Amiri
Bayrem Jemmali
Mohamed Amine Ferchichi
Hajer Jeljeli
Rekik Boulbaba
Abderrahmane Ben Gara
Research to assess the effect of single genes on reproductive traits in bovine species is imperative to elucidate genes' functions and acquire a better perspective of quantitative traits. The present study was undertaken to characterize genetic diversity in the bovine growth hormone (GH) gene in a population of 410 Holstein dairy cows in Tunisia. The analyses were based on single nucleotide polymorphisms, and GH-AluI and GH-MspI detections and genotyping were carried out using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. Data were analyzed using a mixed linear model with the MIXED procedure to reveal the possible effect of GH genotypes on reproductive traits. The frequency data of AluI(L//V) and MspI(+//−) alleles were 87.04//12.96 and 70.06//29.94, respectively. The distribution of the frequency of GH genotypes for LL/LV/VV and (−/−)//(+/−)//(+/+) were 77.75//18.59//3.66 and 15.37//29.13//55.50, respectively. The results of the statistical analyses proved that GH-AluI showed a substantial favorable effect on exanimate traits except for the age at first calving; however, only a suggestive effect of GH-MspI on the calving interval (CI) and the days open (DI) was found. The homozygous LL genotype seemed to be advantageous with respect to the CI and the DI compared with LV and VV genotypes. Heterozygous MspI(+/−) cows tended to have a longer CI and DI than MspI(+/+) and MspI(−/−) cows, but the difference was not statistically significant. A significant effect of different GH-AluI–MspI combined genotypes was found on the number of inseminations per conception, the CI and the DI, and the LL/– combined genotype seemed to be associated with better reproductive performance. Based on these results, the LL genotype of the GH locus can be considered to be a favorable genotype for reproductive traits in Holstein dairy cattle, although these findings need to be confirmed by further research before polymorphisms can be used in a marker-assisted selection program.
- Article
(185 KB) - Full-text XML
- BibTeX
- EndNote
The availability of dense genomic information has increased genome-wide association research for quantitative traits in bovine species. Fertility is considered to be a functional polygenic trait controlled by numerous genetic loci and biased by environmental factors. Hence, linked candidate genes of the encoding loci are searched to optimize reproductive performance.
Growth hormone (GH) belongs to a family of somatolactogenic hormones which include placental lactogen, prolactin and anthers hematopoietic growth factors (Cosman et al., 1990). GH is an anabolic hormone synthesized and secreted by the somatotroph cells, it is a major regulator of postnatal growth and metabolism in mammals, and it plays an important role in lactation, protein, lipid and carbohydrate metabolism, tissue growth, and fertility in cows (Akers, 2006; Lucy, 2008; Thidar et al., 2008). The growth promoting and metabolic actions of GH are generally mediated by the insulin-like growth factor I (IGF-I) (Ramesha et al., 2015).
GH is a polypeptide hormone with a sequence 191 amino acid, its length is approximately 1800 bp and its production is linked to chromosome region 19q26 in the bovine genome (Hediger et al., 1990) with five exons and four introns.
Several polymorphisms of the bovine GH gene have been described and three polymorphic sites have been identified within the fifth exon of this gene (Yao et al., 1996)
Many researchers have studied the associations of GH gene polymorphisms with production traits in dairy or beef cattle (Balogh et al., 2009a; Heidari et al., 2012) owing to the critical role of GH with respect to growth, body composition, metabolism regulation, lactation and mammary gland development (Lagziel and Soller, 1999). A significant relationship between polymorphisms in the bovine GH gene and lactation performance has been reported (Zhou et al., 2005; Balogh et al., 2009b; Mullen et al., 2010).
Lucy et al. (1993) reported a polymorphic site for AluI restriction endonuclease, due to cytosine to guanine transversion at position 2141 which induced a change of the leucine (L) into a valine (V) in the amino acid sequence at position 127 of the protein chain of bovine GH. The genotype of GH AluI polymorphism was reported to be associated with birth weight in dairy cattle (Biswas et al., 2003) and mature live weight in beef cattle (Zwierzchowski et al., 2001). Current knowledge regarding the interrelation between AluI polymorphism and reproduction is limited, and only a few studies have investigated this association.
No significant association has been reported between AluI polymorphism and the following traits: age at the first calving (Kovács et al., 2006; Grossi et al., 2015), the number of oocytes suitable for in vitro maturation, the number of matured oocytes, the mean oocyte diameter, and the number of embryos produced (Lechniak et al., 2002). However, a significant association was observed for milk yield, milk fat and protein (Dybus, 2002; Kovács et al., 2006), in addition to beef bulls non-return rates (Lechniak et al., 2002).
The GH gene was digested with MspI restriction endonuclease; GH-MspI polymorphism is located in intron 3 of the GH gene at position 1547 (Zhang et al., 1993). As a consequence, two alleles occur, and MspI (−) contains a T insertion at the +837 position and a C–G transition at the +837 position (Lee et al., 1994).
To date, several studies have investigated the association of GH-MspI polymorphism with milk and meat production traits (Zhou et al., 2005; Beauchemin et al., 2006; Katoh et al., 2008), but only a few studies have examined its effect on reproductive traits (Unanian et al., 2002; Lechniak et al., 2002; Gorbani et al., 2009).
Based on the abovementioned evidence, the present investigation was carried out to identify GH-AluI and GH-MspI polymorphisms and uncover their association with reproductive traits in Holsteins dairy cattle.
2.1 Experimental design
Four hundred and ten (410) Holstein dairy cows from the Agricultural Development farm in northern Tunisia were included in the present work. These animals were either imported as pregnant heifers from European countries or were born on the farm.
Cows were observed for signs of estrus twice daily, as well as during the morning and afternoon milking. Animals detected in estrus were artificially inseminated. Pregnancy diagnostic by rectal palpation was performed during the 60–70 days after insemination, and pedigree information was available for all cows in the herd.
All cow milk production and reproductive performance data were systematically recorded spanning the time period from January 2013 to December 2017. The reproduction data were derived from the national Office of Livestock and Pasture (OEP) database.
2.2 Blood samples
Five milliliters of venous blood was collected under sterile conditions, from the jugular vein of animal using a 5 mL polypropylene centrifuge tube containing 0.5 mL of 2.7 % EDTA solution as an anticoagulant. The experiment was carried out with the approval of the Institutional Animal Ethics Committee (IAEC). The centrifuge tube was tightly capped and shaken gently to facilitate the thorough mixing of the blood with the anticoagulant. Blood samples were transported to the laboratory in an icebox containing ice packs and stored in a refrigerator at −20 ∘C until the isolation of the DNA was carried out.
2.3 Genotyping
Genomic DNA was isolated from frozen blood using the NucleoSpin Blood kit (innuPREP Blood DNA Mini Kit) following the manufacturer's instructions. The integrity of the DNA was examined using electrophoresis on a 1 % agarose gel, and acceptable samples were used for PCR reaction.
Genotyping was performed using a PCR reaction mixture containing 4 µL of genomic template, 2.5 µL of buffer, 2.5 µL of dNTP, 2.5 µL of each of the primers (F/R), 0.2 µL of the heat-resistant DNA polymerase (Taq polymerase) and 10.8 µL of autoclaved distilled water in a final volume of 25 µL. The primers used were designed to amplify 428 and 329 bp GH gene fragments. Table 1 shows the primers used for amplification.
PCR was carried out with an initial step of 95 ∘C for 5 min, followed by 35 cycles of 95 ∘C for 30 s, 59 ∘C or 60 ∘C for 30 s (for AluI and MspI, respectively), 72 ∘C for 30 s and a final extension of 72 ∘C for 10 min. The PCR amplification yielded 428 and 329 bp fragments that were revealed using electrophoresis on a 2 % agarose gel.
Restriction endonuclease AluI and MspI were used to digest a 428 and 329 bp PCR product, respectively. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) reaction requires 12 µL of the PCR product, 0.2 µL of BSA (bovine albumin serum), 2.5 µL of the reaction buffer, 0.3 µL of the restriction enzyme, 10 µL of autoclaved distilled water in a total volume of 25 µL incubated at 40 ∘C for at least 11 h. The PCR-RFLP fragments were separated using horizontal electrophoresis in 2.5 % agarose gels stained with ethidium bromide and visualized under a UV transilluminator, the gel was documented using Gel Doc equipment, and the genotypes were determined according to band sizes.
Allele and genotype frequencies were calculated using genotyping data from each gene based on the population analyzed. Chi-square statistics (χ2) were used to check whether the populations were consistent with the Hardy–Weinberg equilibrium.
2.4 Reproductive traits
Lactation and several other reproduction traits were used to investigate the effect of the polymorphisms studied on the performance of dairy cows. All data regarding cow milk production and reproductive performance were systematically recorded spanning the time period from January 2013 to December 2017; the following traits were derived from these records: the integrated age at first calving (AGE_1CALV), the calving interval (CI), the number of inseminations per conception (NINS) and the days open (DO; the interval between calving and the next successful insemination). These traits are believed to be the most important and available parameters of reproductive efficiency for the herd. Descriptive statistics for these traits are presented in Table 2.
2.5 Statistical analysis
The statistical analysis was performed by repeated measures using the MIXED procedure in SAS software (SAS 9.0 for Windows). The resulting mixed linear model included the following effect which was used to evaluate the impact of GH polymorphisms on reproduction traits of cows:
where y is the variable dependent on animal, μ is the overall mean, GAlui is the fixed effect of the GH-AluI genotype (LL, LV, VV), GMspj is the fixed effect of the GH-MspI genotype (−/−, +/−, +/+), origink is the fixed effect of the cows' country of origin (k=3), seasonl is the fixed effect of the of calving season (l can represent a value from 1 to 4 which refers to winter: December–February, spring: March–May, summer: June–August, autumn: September–November, respectively), paritym is the fixed effect of the order of parity (m=4), lamenessn is a fixed effect that indicates whether the cow has been diagnosed with lameness or not (n=2) and eijklmn is the random residual error.
The F statistic and the T statistic were used to compare the effect of each factor. A probability of p≤0.05 and p≤0.01 were considered to be statistically significant and highly significant, respectively.
3.1 Gene frequency
The DNA isolated was of a good quality. The 428 and 329 pb fragments from the GH gene were characterized and successfully amplified from the DNA of each sample; this indicated a strong conservation of the DNA sequence existing in cattle.
The χ2 test showed that the allele frequency and genotypes of GH-AluI and GH-MspI were in Hardy–Weinberg equilibrium.
Visualization under UV light revealed that GH-AluI exhibited three band patterns. Fragments that represent the leucine variant (LL) gave rise to the 265//96//51 pb band, AluI fragments gave rise to the 265//147 pb band that represented the valine variant (VV), and the (LV) pattern gave rise to the 265//147//96//51 pb band.
The following DNA restriction fragments were also obtained as a result of MspI restriction: 329 bp for the MspI(−/−) genotype, 329//224//105 bp for the MspI(+/−) genotype and 224//105 bp for the MspI(+/+) genotype.
Table 3 shows genotypic and allelic frequencies of GH-AluI and GH-MspI polymorphisms in the herd studied. GH-AluI genotype frequencies were 77.75, 18.59 and 3.66 for LL, LV and VV, respectively. These results showed that the L allele was significantly more frequent than the V allele (87.045 vs. 12.955).
The GH-MspI frequency distribution showed that the MspI (+/+) genotype was the most abundantly found (55.50), followed by MspI (+/−) (29.13). The least frequently found genotype was MspI (−/−) (15.37). The MspI (+) allele was more frequent than the MspI (−) allele (70.065 vs. 29.935).
3.2 Factors affecting reproduction traits
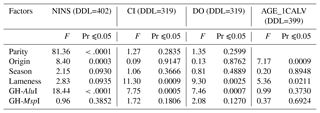
The investigation of the effect of non-genetic factors on reproduction traits showed that parity only had a significant effect on the NINS (p<0.0001) but had no effect on the CI or the DO. The cows' country of origin significantly affected the NINS (p=0.0003) and the AGE_1CALV (p=0.0009) but had no effect on the CI or the DO. The diagnosis of lameness had a highly significant effect on the CI (p=0.0009) and the DO (p=0.0025), a significant effect on the AGE_1CALV (p=0.0211) and no effect on the NINS. The calving season had no impact on any of the traits analyzed.
As shown in Table 4, genetic polymorphisms were found to be related to the abovementioned traits. The GH-AluI significantly affected the NINS (p<0.0001), the CI (p=0.0005) and the DO (p=0.0007) but did not affect the AGE_1CALV. However, the GH-MspI had no effect on any of the traits analyzed.
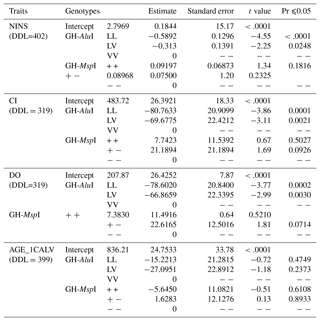
Table 4The effect of sources of variation on reproductive traits. DDL refers to degrees of freedom, and Pr refers to probability.
The effect of genetic polymorphisms on reproductive traits was also compared to the GH-AluI and GH-MspI variants (Table 5). The replacement of the L allele by the V allele was found to lead to an increase in the NINS, the CI and the DO, and the performance of the LL genotype was significantly higher compared with similar values of the VV and LV genotypes. Cows with the LL genotype had the lowest NINS (−0.5892) compared with cows with VV and LV genotypes – the difference was highly significant (p<0.0001).
The GH-AluI polymorphism had a substantial effect on the CI (p<0.0001), and the LL genotype shows a shorter CI (−80.76 days) compared with the VV genotype. The difference between the LV and VV genotypes was also considerable (p=0.0021) with a shorter CI for the LV genotype (−69.68 days). The statistical model that estimates the genotype effect revealed that the LL and LV genotypes were significantly associated with the DO (p=0.0002 and p=0.0030, respectively), and that LL and LV cows tend to conceive 78.6 and 66.87 days before VV cows, respectively. There was no effect of the substitution of the L allele by the V allele on the AGE_1CALV: VV cows tended to have their first calving 15.22 and 27.10 days later than LV and LL cows, respectively, but the differences were not statistically significant.
There was no detectable significant effect of the GH-MspI on the traits analyzed, although polymorphism had a suggestive effect on the CI (p=0.0926) and the DO (p=0.0714). MspI (+/−) cows tended to have the longest CI and DO, but the difference was still not significant.
Table 5Association of GH-AluI and GH-MspI genotypes with reproduction trait. DDL refers to degrees of freedom, and Pr refers to probability.
A statistical evaluation of the association of the GH-AluI–MspI combined genotypes on reproductive traits showed a significant effect on the NINS (p<0.0001), the CI (p=0.0024) and the DO (p=0.0022), although no effect was found on the AGE_1CALV (Table 6).
The estimate of the VV/− genotype intercept for the NINS, the CI, the DO and the AGE_1CALV is 2.610, 498.45, 218.79 and 812.61, respectively. However, differences in the combined genotype performance is only significant for the CI and the DO.
There were no genetic factors (namely parity, origin, calving season and lameness) that clearly accounted for the reproduction traits examined in this study.
Parity showed a significant relationship with the NINS, with primiparous cows requiring a lower number of inseminations per conception than pluriparous cows. The impact of the cow's origin with respect to the NINS and the AGE_1CALV provided evidence that exported cows had better reproductive performance than native cows in this study. Regarding the antagonist correlation between lameness and reproduction traits, cows that were diagnosed with lameness did not perform as well as cows that were not diagnosed. In addition, differences in the feeding and climatic features of the calving season had no effect on the traits analyzed except for the fact that the NINS was slightly influenced by calving season.
The majority of cows were homozygous for the leucine allele (L; 77.75 %); only 3.66 were homozygous for the valine allele (V), whilst 18.59 % were heterozygous. Therefore the frequency of the V allele was lower than the L allele (87.045 vs. 12.955). Allele and genotype frequencies for the GH-AluI polymorphisms given in this work were in agreement with those reported in previous studies, which have been discussed extensively in Holstein dairy cows (Balogh et al., 2009b; Ruprechte et al., 2011; Grossi et al., 2015).
However, other findings have shown the dominance of the heterozygous genotype LV and the absence of the homozygous genotype VV (Aruna et al., 2004; Hadi et al., 2015).
The most frequent genotypes for GH-MspI were homozygous MspI(+/+) (55.50), whereas the homozygous MspI(−/−) genotype was the least frequent (15.37). Hence, the dominance of the Msp(+) allele was evident compared with the MspI(−) allele (70 065 vs. 29 935). Results of the present work are in agreement with those published by Dybus et al. (2002) in Polish Black-and-White cattle and by Zhou et al. (2006) in China Holstein cows. However, Hartatik et al. (2018) reported a higher frequency of the MspI(+/−) genotype than the MspI(−/−) and MspI(+/+) genotypes in Pesisir cattle and cross breeds, in addition to the fact that MspI(−) allele was more frequent than the MspI(+) allele (0.533 vs. 0.467).
There are only a few studies that have investigated the relationship between GH-AluI and GH-MspI polymorphisms and reproductive traits in Holstein dairy cattle. Evaluations of this relationship in this work showed a significant effect of the GH-AluI on the NINS, the CI and the DO but no effect on the AGE_1CALV. Furthermore, no significant association between the GH genotype and the interval from calving to first ovulation was observed by Balogh et al. (2009b).
Similarly, no obvious association of GH with AGE_1CALV and CI was observed in Canchim and Hungarian Holstein–Friesian cattle (Kovacs et al., 2006; Grossi et al., 2015) and with AGE_1CALV in a Hungarian Holstein–Friesian bull dam population (Katalin et al., 2006). Moreover, Kovacs et al. (2006) reported no noteworthy effect of GH-AluI on the CI, the number of calves or the dry period in a Hungarian Holstein–Friesian bull dam. Ruprechter et al. (2011) found no effect of the GH genotype on the calving–first service interval, the number of services per conception or the total pregnancy rate, and no other significant effects were observed with respect to the number of matured oocytes or the number of embryos produced (Lechniak et al., 2002).
In our study, the LL genotype is considered the most favorable with respect to reproductive traits, as cows carrying the LL genotype showed better performance than LV and VV cows – this difference was highly significant for NINS, the CI and the DO. Lechniak et al. (1999) reported no significant genotype effect for lower ejaculate volumes for leucine homozygous bulls and greater non-return rates of VV beef bulls at 60 days postpartum.
Sabour et al. (l997) reported that V/V genotypes were related to better milk traits, especially higher milk protein compared with the other genotypes. These findings are in line with our result if we consider the established antagonistic correlation between reproduction and the milk production function, which means that the GH genotype may directly or indirectly affect reproduction performance. In addition, numerous previous studies have suggested an additive effect of the L allele on milk production and contents with LL cows reportedly yielding more milk, fat and protein than LV and VV cows (Lucy et al., 1991; Shariflou et al., 2000; Dybus, 2002).
This examination of the impact of the GH locus on reproductive traits showed that GH-MspI polymorphism only had a suggestive, non-significant effect on the CI and the DO. To date, few studies have investigated the effect of GH-MspI polymorphism on reproductive traits of bulls, and the results of these studies have reported a significant association between GH-MspI polymorphisms and the following sperm quality traits in bulls: fresh sperm motility, sperm concentration, minor and major defects, scrotal circumference and testicular growth after puberty (Lechniak et al., 2002; Unanian et al., 2002; Gorbani et al., 2009); the reproductive traits of beef cattle have also been studied (Anggraeni et al., 2017).
Arango et al. (2014) reported that MspI polymorphism is associated with age at first service, first birth, first postpartum service and second birth. Furthermore, the MspI (+/+) genotype had the greatest effect on the age at first service and was considered the optimal genotype for reducing age at first service, first birth, first postpartum service and second birth. In addition, several inconsistent results have reported the effect of GH-MspI polymorphism on growth and milk production traits (Dybus et al., 2004; Pereira et al., 2005; Zhou et al., 2006; Katoh et al., 2008). MspI(+/+) cows are considered to yielded more milk, fat and protein than +/− individuals (Zhou et al., 2006).
Significant differences between cows with different GH-AluI–MspI combined genotypes were found at the 5 % level for the CI; cows carrying the LL/− genotype showed significantly (p=0.021) better performance, and their CI was 95.754 days lower than VV/− cows. The GH-MspI–AluI combined genotypes also significantly affected the NINS (P<0.0001); cows with the VV/ genotype needed the highest NINS in the herd, although this tendency was not significant.
The effect of the combined genotypes was also notable with respect to the DO (p=0.0022), further confirming the difference between cows of different genotypes. The LL/− cows were significantly (p=0.0298) associated with the shortest DO compared to the estimated intercept of the VV/− genotype. However, no significant association was found between cows with different GH-AluI–MspI combined genotypes and the AGE_1CALV.
As previously stated, the Alu/LL genotype showed the best relationship with reproductive traits, whereas the GH-MspI only had a suggestive effect on the CI and the DO. Hence, the LL/− combined genotype can be expected to be associated with better performance. Very few studies have investigated the effect of GH-AluI–MspI combined genotypes in dairy cattle. However Dybuset al. (2004) reported that VV/ cows produced milk with the highest fat and protein content.
In this study, AluI and MspI polymorphisms of the bovine GH gene were identified in a population of Holstein dairy cows in Tunisia which displayed L allele and MspI(+) allele dominance. GH-AluI significantly affected the NINS, the CI and the DO; however GH-MspI had no effect on any of the traits evaluated except for a slight tendency on the CI and the DO. GH-MspI–AluI combined genotypes had a highly significant affect on the NINS, the CI and the DO, and cows with the LL/− genotype tended to have better reproductive traits.
Under field conditions, the detection of polymorphisms in genes related to reproductive traits and the identification of the allele which results in a phenotype of interest can be considered as candidate markers of reproductive traits for cattle breeders.
The data are not publicly accessible; they are the property of the National Livestock and Pasturing Office (OEP) of Tunisia.
The authors declare that they have no conflict of interest.
The National Livestock and Pasturing Office (OEP) are gratefully acknowledged for
their contribution to this work and for making their database available. The authors are also grateful
to the Chergui Society of Agricultural Development
and their personnel for their kind and helpful contribution to this work.
Edited by: Steffen Maak
Reviewed by: Ghada Nouairia
Akers, R. M.: Major advances associated with hormone and growth factor regulation of mammary growth and lactation in dairy cows, J. Dairy Sci., 89, 1222–1234, 2006.
Anggraeni, A., Talib, C., Asmarasari, S. A., Herawati, T., and Andreas, E.: Genetic Polymorphisms of IGF1, GH, and OPN Genes in Crosses Peranakan Ongole Cattle Based on Birth Type in Central Java, JITV, 22, 165–172, 2017.
Arango, J. G., Echeverri, Z. J., and López, H. A.: Association of the bovine growth hormone gene with Holstein cattle reproductive parameters, Rev. MVZ Córdoba 19, 4249–4258, 2014.
Aruna, P., Chakravarty, A. K., Bhattacharya, T. K., Joshi, B. K., and Arjava, S.: Detection of Polymorphism of Growth Hormone Gene for the Analysis of Relationship between Allele Type and Growth Traits in Karan Fries Cattle, Asian-Aust, J. Anim. Sci. 17, 1334–1337, 2004.
Balogh, O., Kovács, K., Kulcsár, M., Gáspárdy, A., Fébel, H., Zsolnai, A., Fésus, C., Delavaud, C., Chilliard, Y., Gilbert, R., and Huszenicza, G.: Interrelationship of growth hormone AluI polymorphism and hyperketonemia with plasma hormones and metabolites in the beginning of lactation in dairy cows, Livest. Sci., 123, 180–186, 2009a.
Balogh, O., Kovacs, K., Kulcsar, M., Gaspardy, A., Zsolnai, A., Katai, L., Pecsi, A., Fesus, L., Butler, W. R., and Huszenicza, G.: AluI polymorphism of the bovine growth hormone (GH) gene, resumption of ovarian cyclicity, milk production and loss of body condition at the onset of lactation in dairy cows, Theriogenology, 71, 553–559, 2009b.
Beauchemin, V. R., Thomas, M. G., Franke, D. E., and Silver, G. A.: Evaluation of DNA polymorphisms involving growth hormone relative to growth and carcass characteristics in Brahman steers, Gene. Mol. Res., 5, 438–447, 2006.
Biswas, T. K., Bhattacharya, T. K., Narayan, A. D., Badola, S., Kumar, P., and Sharma A.: Growth hormone gene polymorphism and its effect on birth weight in cattle and,buffalo, Asian-Aust. J. Anim. Sci., 16, 494–497, 2003.
Cosman, D., Lyman, S. D., Idzerda, R. L., Beckmann, M. P., Park, L. S., Goodwin, G., and March, C. J.: A new cytokine receptor superfamily, Trends Biochem. Sci., 15, 265, 1990.
Dybus, A.: Associations between Leu/Val polymorphism of growth hormone gene and milk production traits in Black and White cattle, Arch. Tierz., 45, 421–428, 2002.
Dybus, A., Grzesiak, W., Szatkowska, I., and Błaszczyk, P.: Association between the growth hormone combined genotypes and dairy traits in Polish Black-and-White cows, Anim. Sci. Pap. Rep., 22, 185–194, 2004.
Gorbani, A., Vaez Torshizi, R., Bonyadi, M., and Amirinia, C.: A MspI PCR-RFLP within bovin growth hormone gene and its association with sperm quality traits in Iranian Holstein bulls, Afr. J. Biotechnol., 8, 4811–4816, 2009.
Grossi, D. A., Buzanskas, M. E., Grupioni, N. V., Paz, C. C. P., Regitano, L. C. A., Alencar, M. M., Schenkel, F. S., and Munari, D. P.: Effect of IGF1, GH, and PIT1 markers on the genetic parameters of growth and reproduction traits in Canchim cattle, Mol. Biol. Rep., 42, 245–251, https://doi.org/10.1007/s11033-014-3767-4, 2015.
Hadi, Z., Atashi, H., Dadpasand, M., Derakhshandeh, A., and Ghahramani, M. M.: The relationship between growth hormone polymorphism and growth hormone receptor genes with milk yield and reproductive performance in Holstein dairy cows, Iran J. Vet. Res. Summer, 16, 244–248, 2015.
Hartatik, T., Putra, D. E., Volkandari, S. D., Kanazawa, T., and Sumadi: Genotype analysis of partial growth hormone gene (GH891|MspI) in Pesisir cattle and Simmental-Pesisir crossbred cattle, J. Indonesian Trop. Anim. Agric., 43, 1–8, 2018.
Hediger, R., Johnson, S. E., Barendse, W., Drinkwater, R. E., Moore, S. S., and Hetzel, J.: Assignment of the GH gene locus to 19q26qter in cattle and to 11q25qter in sheep by in situ hybridization, Genomics, 8, 171–174, 1990.
Heidari, M., Azari, M. A., Hasani, S., Khanahmadi, A., and Zerehdaran, S: Effect of Polymorphic Variants of GH, Pit-1, and β-LG Genes on Milk Production of Holstein Cows, Russ. J. Genet., 48, 417–421, 2012.
Katalin, K., József, V., Attila, Z., István, G., and László, F.: Associations between the AluI polymorphism of growth hormone gene and production and reproduction traits in a Hungarian Holstein- Friesian bull dam population, Arch. Tierz., 49, 236–249, 2006.
Katoh, K., Kouno, S., Okazaki, A., Suzuki, K., and Obara, Y.: Interaction of GH polymorphism with body weight and endocrine functions in Japanese black calves, Domest. Anim. Endocrinol., 34, 25–30, 2008.
Kovács, K,. Völgyi-Csík, J., Zsolnai, A., Györkös, I., and Fésüs, L.: Associations between the AluI polymorphism of growth hormone gene and production and reproduction traits in a Hungarian Holstein-Friesian bull dam population, Arch. Tierz., 49, 236–249, 2006.
Lagziel, A. and Soller, M.: DNA sequences of SSCP haplotypes at the bovine growth hormone (bGH) gene, Anim. Genet., 30, 362–365, 1999.
Lechniak, D., Machnik, G., Szydlowski, M., and Switonski, M: Growth hormone gene polymorphism and reproductive performance of AI bulls, Theriogenology, 52, 1145–1152, 1999.
Lechniak, D., Adamowicz, T., Stanislawski, D., and Kaczmarek, D.: In vitro maturation and fertilization of bovine oocytes in relation to GH gene polymorphism (Leu/Val), Reprod. Nutr. Dev., 42, 275–80, 2002.
Lee, B. K., Crooker, B. A., Hansen, L. B., and Chester-Jones, H.: Polymorphism in the third intron of somatotropin (bST) gene and its association with selection for milk yield in Holstein cows, J. Anim. Sci., 72, 316, 1994.
Lucy, M. C.: Functional differences in the growth hormone and insulin-like growth factor axis in cattle and pigs: implications for post-partum nutrition and reproduction, Reprod. Domest. Anim., 43, 31–39, 2008.
Lucy, M. C., Hauser, S. D., Eppard, P. J., Krivi, G. G., and Collier, R. J.: Genetic polymorphism within the bovine somatotropin (bST) gene detected by polymerase chain reaction and endonuclease digestion, J. Dairy Sci., 74, 284, 1991.
Lucy, M. C., Hauser, S. D., Eppard, P. J., Krivi, G. G., Clark, J. H., Bauman, D. E., and Collier, R. J.: Variants of somatotropin in cattle: gene frequencies in major dairy breeds and associated milk production, Domest. Anim. Endocrin., 10, 325–333, 1993.
Mullen, M., Berry, D., Howard, D., Diskin, M., Lynch, C., Berkowicz, E., Magee, D., MacHugh, D., and Waters, S.: Associations between novel single nucleotide polymorphisms in the Bos taurus growth hormone gene and performance traits in Holstein-Friesian dairy cattle, J. Dairy Sci., 93, 5959–5969, 2010.
Pereira, A. P., De Alencar, M. M., De Oliveira, H. N., and De Regitano, L. C.: Association of GH and IGF-1 polymorphisms with growth traits in a synthetic beef cattle breed, Genet. Mol. Biol., 28, 230–236, 2005.
Ramesha, K. P., Rao, A., Basavaraju, M., Geetha, G. R., Kataktalware, M. A., and Jeyakumar, S.: Genetic variability of bovine GHR, IGF-1 and IGFBP-3 genes in Indian cattle and buffalo, S. Afr. J. Anim. Sci., 45, https://doi.org/10.4314/sajas.v45i5.5, 2015.
Ruprechter, G., Carriquiry, M., Ramos, J. M., Pereira, I., and Ana, M.: Metabolic and endocrine profiles and reproductive parameters in dairy cows under grazing conditions: effect of polymorphisms in somatotropic axis genes, Acta Vet. Scand., 53, https://doi.org/10.1186/1751-0147-53-35, 2011.
Sabour, M. P., Lin, C. Y., and Smith, C.: Association of genetic variants of bovine growth hormone with milk production traits in Holstein cattle, J. Anim. Breed Genet., 114, 435–42, 1997.
Shariflou, M. R., Moran, C., and Nicholas, F. W.: Association of the Leu variant of the bovine growth hormone (bGH) gene with increased yield of milk, fat, and protein in Australian Holstein-Friesians, Aust. J. Agr. Res., 51, 515–522, 2000.
Thidar, M., Yoshida, H., Ito, H., He, T., Inoue, M., and Kuwayama, H.: Combined administration of ghrelin and GHRH synergistically stimulates GH release in Holstein preweaning calves, Domest. Anim. Endocrin., 34, 118–123, 2008.
Unanian, M. M., Barreto, C. C., Cordeiro, C. M. T., Freitas, A. R., and Josahkian, L. A.: Possible association between bovine growth hormone gene polymorphism and reproductive traits, Braz. Arch. Boil. Tech., 45, 293–299, 2002.
Yao, J., Aggrey, S. E., Zadworny, D., Hayes, J. F., and Kuhnlein, U.: Sequence variations in the bovine growth hormone gene characterized by single-strand conformation polymorphism (SSCP) analysis and their association with milk production traits in Holsteins, Genetics, 144, 1809–1816, 1996.
Zhang, H. M., Maddock, K. C., Brown, D. R., Denise, S. K., and Ax, R. L.: A novel of the bovine somatotropin gene detected by PCR-RFLP analysis, Anim. Sci., 71, 2276, 1993.
Zhou, G. L., Jin, H. G., Liu, C., Guo, S., Zhu, Q., and Wu, Y. H.: Association of genetic polymorphism in GH gene with milk production traits in Beijing Holstein cows, J. Biosci., 30, 595–598, 2005.
Zhou, G. L., Zhu, Q., Jin, H. G., Liu, C., and Guo, S.: Genetic Variation of Growth Hormone Gene and Its Relationshipwith Milk Production Traits in China Holstein CowsAsian-Aust, J. Anim. Sci., 19, 315–318, 2006.
Zwierzchowski, L., Oprzadek, J., Dymnicki, E., and Dzierzbicki, P.: An association of growth hormone, kappa-casein, betalactoglobulin, leptin and pit-1 loci polymorphism with growth rate and carcass traits in beef cattle, Anim. Sci. Pap. Rep., 19, 65–77, 2001.