the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Detection of insertion/deletions (indels) of the ATBF1 gene and their effects on growth-related traits in three indigenous goat breeds
Zhenyu Wei
Ke Wang
Yiqing Hui
Hailong Yan
Haijing Zhu
Lei Qu
Chuanying Pan
Hong Chen
Xianyong Lan
The AT motif-binding factor (ATBF1), also known as zinc finger homeobox 3 (ZFXH3), is necessary for activating the POU1F1 gene; thus, the ATBF1 gene greatly affects the grow traits in animals. The objective of this work was to explore novel indel (insertion/deletion) variations and their associations with growth traits in three native Chinese goat breeds. Two indels within the ATBF1 gene were found in the Shaanbei white cashmere goat (SWCG; n=581), the Guanzhong dairy goat (GZDG; n=334) and the Hainan black goat (HNBG; n=270) for the first time using 12 pairs of primers. Association analysis revealed that the P1-12-bp indel was consistently correlated with the body height of the three breeds, and individuals with ID (insertion/deletion) and DD (deletion/deletion) genotypes had a higher body weight than the II (insertion/insertion) genotype (P=0.036); the P11-6-bp indel was consistently correlated with chest circumference and hip width of the three breeds. Moreover, these two loci were associated with other several growth-related traits in different breeds. Hence, these findings indicated that the goat ATBF1 gene had marked effects on growth traits and the growth-trait-related loci, which would contribute to improving the growth-related traits of local breeds in the goat industry by implementing marker-assisted selection (MAS).
- Article
(1461 KB) - Full-text XML
- BibTeX
- EndNote
The demand for goat meat has increased quickly in many countries, especially China. Increasing profitability in goat rearing requires that certain growth-related traits of local goat breeds should be significantly improved (Zhao et al., 2014). Genetic variation in growth potentials is large among populations and commonly adequate to support reasonable rates of genetic improvement within populations. An effective DNA marker-assisted selection (MAS) would speed up the development and improvement goat products. Consequently, it is meaningful to screen some important genes and explore their polymorphism with growth-related traits.
Table 1PCR primer sequences of the goat ZFXH3 gene for amplification.
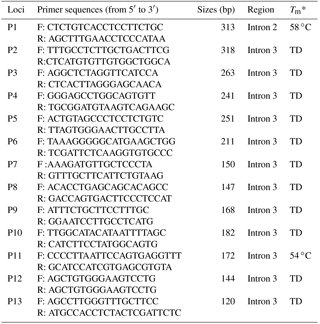
Note: F: forward; R: reverse; Tm*: temperature at half of the maximum value of UV absorption during the thermal denaturation of the DNA double-helix structure. TD: touchdown PCR protocol; anneal temperature is during 68 to 50 ∘C.
The AT motif-binding factor (ATBF1), also known as zinc finger homeobox 3 (ZFHX3), is a crucial cancer suppressor gene. It is worth mentioning that ATBF1 could promote the expression levels of the growth hormone prolactin and TSH-β by binding to the enhancers of POU1F1 (also known as Pit1) and PROP1 (Darling et al., 1998; Qi et al., 2008; Araujo et al., 2013; Yan et al., 2018). These studies showed that the ATBF1 gene was hypothesized to produce important effects on cell differentiation and early development, suggesting that the ATBF1 gene would affect the growth traits in animals. In our team, Zhang et al. (2015) first explored the single nucleotide polymorphisms (SNPs) and haplotype structure of the goat ATBF1 gene and analyzed its associations with growth-related traits in two Chinese goat breeds. Xu et al. (2017) identified crucial SNP variations within the ATBF1 gene in native Chinese cattle breeds. These functional experiments suggested that the ATBF1 gene would affect the growth traits of livestock.
As another type of DNA marker, indel (insertion/deletion) can be effectively used in MAS. Few studies about the indel of the goat ATBF1 gene and its effects on growth traits have been reported. Hence, in this study we explored the novel indel loci and analyzed its associations with growth-related traits to uncover the relationship between the indel loci of the ATBF1 gene and growth-related traits in three Chinese goat breeds. These findings might provide molecular markers for MAS programs to improve the growth-related traits of local breeds in the goat industry.
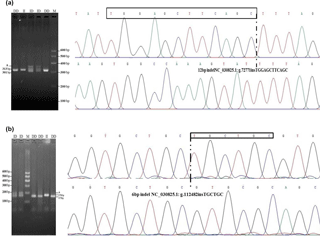
Figure 1Agarose gel electrophoresis patterns and sequencing maps for the ATBF1 gene P1-12-bp indel and P11-6-bp indel. (a) For the P1-12-bp, PCR products showed three genotypes at this locus where the insertion type (II genotype) consisted of 313 bp, deletion type (DD) consisted of 301 bp, and the heterozygote type (ID) showed 313 and 301 bp, which accounted for 3.5 % in agarose gel electrophoresis. Sequencing maps showed the homozygotic insertion type (II) and homozygotic deletion type (DD) of P1-12-bp. (b) For the P11-6-bp indel, PCR products showed three genotypes at this locus: the insertion type (II genotype, 178 bp), the deletion type (DD, 172 bp), and the heterozygote type (ID, 178/172 bp), which accounted for 3.5 % in agarose gel electrophoresis. Sequencing maps showed the homozygotic insertion type (II) and the homozygotic deletion type (DD) of the P11-6-bp indel.
2.1 Animals and data collection
Experimental animal samples used in this study were approved under contract by the Review Committee for the Use of Animal Subjects of Northwest A&F University. The care and use of experimental animals was performed in agreement with the ethics commission's guidelines.
A total of 1185 ear samples were collected from healthy adult goats belonging to three characteristic Chinese native goat breeds: the Shaanbei white cashmere goat (SWCG; n=581), the Guanzhong dairy goat (GZDG; n=334) and the Hainan black goat (HNBG; n=270). All SWCG individuals' genomic DNA were extracted from the ear tissue of animals reared at the SWCG goat breeding farm in Yulin, Shaanxi Province, and for which there were records of growth traits data (Cui et al., 2018; Wang et al., 2017). In particular, 209 of 581 SWCGs with body weight and growth trait data were collected at the SWCG goat breeding farm in Yulin, Shaanxi Province, on 3–5 April 2018 and used to explore the relationships between these indel loci and body weight. GZDG individuals were collected from the breeding farm in Fuping, Shaanxi Province. The Hainan black goats were fed in Hainan Province. The growth traits data of GZDG and HNBG were collected by the staff at these farms and included body height (BH), body length (BL), chest width (CW), chest depth (CD), chest circumference (CC), height across the hip (HAH), rump length (RL), hip width (HW) and circumference of the cannon bone (CCB) (Zhang et al., 2015; Yan et al., 2018).
2.2 DNA isolation and genomic DNA pools construction
Genomic DNA samples were extracted using a high salt-extraction method (Aljanabi and Salah, 1997); all genomic DNA samples were diluted to the working concentration of 20 ng µL−1 for the DNA pool construction and polymerase chain reaction (PCR) amplification and stored at −20 ∘C. DNA pools comprised 30 randomly selected individual genomic DNA samples from the three breeds (Wang et al., 2018). The three DNA pools were used as templates for PCR amplification, and the product of amplification was used to sequence and explore indels in the ATBF1 gene.
2.3 Primers design and DNA sequencing
Information about ZFHX3 indels was obtained from NCBI-dbSNP (https://www.ncbi.nlm.nih.gov/snp; last access: 18 January 2009). To expose indels in the goat ATBF1 gene, 13 pairs of primer were designed using the Primer Premier 5 software based on the goat ATBF1 gene sequence (GenBank Accession No. NC_030825.1) (Table 1). PCR reactions were performed in 25 µL volumes, and the same touchdown PCR protocol was followed as in Wang et al. (2018). The products were sequenced only when they had a single objective band of each pair of primers. Direct sequencing of PCR products was performed using an Applied Bio-systems 3730XL DNA sequencer (Tsingke Beijing, China). Indel variations were identified by the sequence alignment of the sequenced reads against the reference sequence by using the BioXM 2.6 software (NJAU, Nanjing, China).
Table 2Genotypic and allelic frequencies and population indexes for the ATBF1 gene P1-12-bp indel in three Chinese goat breeds.
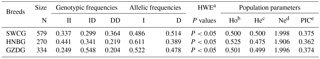
Note: SWCG – Shaanbei white cashmere goat; HNBG – Hainan black goat; GZDG – Guanzhong dairy goat; a HWE – Hardy–Weinberg equilibrium. b Ho: observed homozygosity. c He: heterozygosity. d Ne: effective allele numbers. e PIC: polymorphism information content.
Table 3Genotypic and allelic frequencies and population indexes for the ATBF1 gene P11-6-bp indel in three Chinese goat breeds.
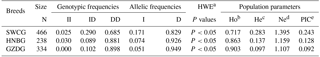
Note: SWCG – Shaanbei white cashmere goat; HNBG – Hainan black goat; GZDG – Guanzhong dairy goat; a HWE – Hardy–Weinberg equilibrium. b Ho: observed homozygosity. c He: heterozygosity. d Ne: effective allele numbers. e PIC: polymorphism information content.
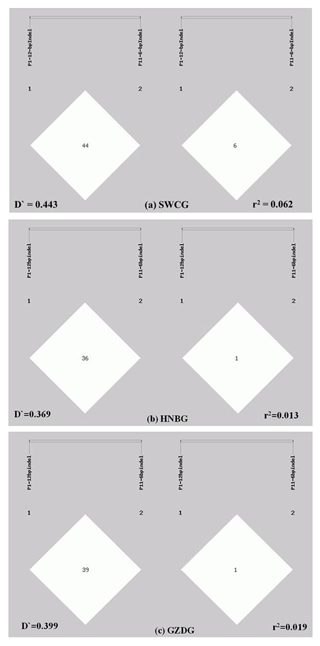
Figure 2Linkage disequilibrium analyses of the P1-12-bp indel and P11-6-bp indel. (a) analysis result in SWCG (; r2=0.062); (b) analysis result in HNBG (; r2=0.013); (c) analysis result in GZDG (; r2=0.019).
2.3.1 Genotyping using PCR-based amplification of fragment length polymorphism
In a previous step, two novel indels were taken out by direct sequencing within the ATBF1 gene, namely NC_030825.1: g.72771insTGGAGCTTCAGC (P1-12-bp indel) and NC_030825.1: g.112482insTGCTGC (P11-6-bp indel). PCR-based agarose gel electrophoresis amplification of fragment length polymorphism was used to genotype these two indels in every individual (n=1185). The 12.5 µL reaction mixture included 0.5 µL of 20 ng µL−1 genomic DNA, 0.5 µL of each primer (10 µM) and 6.25 µL 2× Taq Master mix (BioLinker, Shanghai, China). The amplification protocol included an initial denaturation step at 95 ∘C for 5 min, followed by 32 cycles of 94 ∘C for 30 s, 56 ∘C for 30 s, 72 ∘C for 20 s and a final extension at 72 ∘C for 10 min. The amplification product was analyzed using 3.5 % agarose gel electrophoresis in 0.5×TBE (tris–borate–EDTA buffer) mixed with nucleic acid stain (BBI, Shanghai, China).
2.4 Statistical analysis
Genotypic frequencies and allelic frequencies were calculated according to Botstein's method (Botstein et al., 1980; Falush et al., 2003). The Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) structure of the two loci in SWCG, HNBG and GZDG were analyzed by the SHEsis program (http://analysis.bio-x.cn, last access: 26 March 2018) (Shi and He, 2005; Li et al., 2009). Population parameters, homozygosity (Ho), effective allele number (Ne) and polymorphism information content (PIC) were computed following Nei's methods (Nei, 1973) and performed using PopGene version 1.3.1 (Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, AB, Canada). The associations of the genetic variations and growth-related traits were calculated by Prism 6th (GraphPad software). Multivariate linear regression between body weight and growth traits was calculated by SPSS 19.0 (IBM software).
The statistical linear model was as follows: , where Yijk is the observation of the growth-related traits, μ is the overall mean of each trait, Ai is the fixed effect of age, Gj is the fixed effect of genotype or combined genotype, and eijk is the random residual error (Wang et al., 2017; Yang et al., 2017). The mixed statistics of the linear model analysis did not include the effects of sex, farm, age of dam and sire, or season of birth, which had no significant effects on the variation of traits in the mammal populations (Zhao et al., 2013). The fixed effect of genotypes and age was a major source of variation and the p value for the difference between the least squares means was less than 0.05 (Zhang et al., 2015).
3.1 Indel identification and genotyping of the ATBF1 gene in goat
After DNA sequencing and alignment analysis, one 12-bp indel (NC_030825.1: g.72771insTGGAGCTTCAGC, P1-12-bp indel) and one 6-bp indel (NC_030825.1: g.112482insTGCTGC, P11-6-bp indel) in intron 3 of the ATBF1 gene was identified in all three breeds (Fig. 1). PCR-based amplification of fragment length polymorphism revealed that P1-12-bp and P11-6-bp indel loci each generated three genotypes.
3.2 Genetic parameter analysis of the P1-12-bp indel and P11-6-bp indel
For the P1-12-bp indel, the frequencies of these genotypes were close to each other, and allelic frequencies of I and D were on an approximately equal footing in 1185 individuals (Table 2). The genotype distributions in all these three breeds were not in line with HWE (P<0.05). The PIC value showed that it had medium genetic diversity (). These results suggest that the P1-12-bp indel is polymorphic in these breeds.
For the P11-6-bp indel, the II genotype showed an extreme low frequency; the frequency of the DD genotype was higher than that of ID in the analyzed breeds (Table 3). In particular, the I allele had a lower frequency than D, suggesting that the D allele had a dominant hierarchy. The genotype distribution was not in line with HWE (P<0.05). The PIC value showed that it had low genetic diversity ()
3.3 Linkage disequilibrium analysis of the P1-12-bp indel and P11-6-bp indel
The LD of two indels was analyzed in a population composed of three goat breeds. As shown in Fig. 2, the D′ and r2 values of all three breeds were very low (; r2<0.2). These values showed that the P1-12-bp indel and P11-6-bp indel are incompletely linked. The II genotype of the P11-6-bp indel had an extreme low frequency in all three breeds; this might be one of the possible reasons.
3.4 Relationships between the indel loci and related-growth traits
The associations of the indel loci with growth-related traits were determined. In Table 4, analysis shows that the P1-12-bp indel was obviously correlated with the body height of all three breeds. The DD and ID genotypes had demonstrated significantly superior growth-related traits than genotype II in SWCG (body height, height across the hip), GZDG (body height, height across the hip) and HNBG (body height, body length, chest width, chest depth, chest circumference, hip width and circumference of cannon bone). For body weight, Fig. 3a shows that individuals with ID and DD genotypes in P1-12-bp had a significantly higher body weight than the II genotype (P=0.036).
Table 4Association of the indel within the ATBF1 gene and growth traits of the P1-12-bp indel of three Chinese goat breeds (mean ± SE).
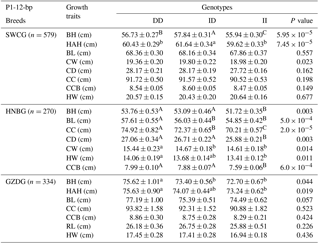
Note: body height, BH; body length, BL; chest width, CW; chest depth, CD; chest circumference, CC; height across the hip, HAH; rump length, RL; hip width, HW; circumference of cannon bone, CCB (a, b, c – P<0.05; A, B, C – P<0.01).
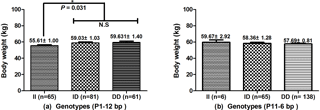
Figure 3Association of the P1-12-bp and P11-6-bp indel with body weight in SWCG. (a) Individuals with ID and DD genotypes have significantly higher body weight than the II genotype in the P1-12-bp indel (P=0.036). (b) There is no significant difference between the P11-6-bp indel and body weight in SWCG. Data represent means ± SE. N.S – not significant. *: P<0.05.
The P11-6-bp indel was obviously correlated with chest circumference and hip width of all three breeds, while there were less than three genotype II individuals in GZDG (Table 5). Genotype DD was found to have significantly superior CC, CCB, BL and HW in SWCG and BH, BL, CC, CD, CW, HW traits in HNBG when compared with genotype ID, while genotype ID had significantly superior CCB, RL, HW traits in GZDG. Figure 3b indicates that there was no significant relation between the P11-6-bp indel and body weight in SWCG.
Table 5Association of the indel within the ATBF1 gene and growth traits of the P11-6-bp indel of three Chinese goat breeds (mean ± SE).
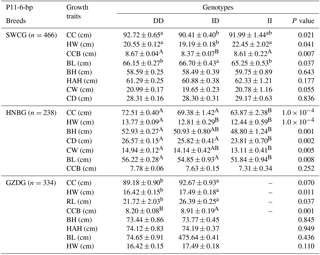
Note: body height, BH; body length, BL; chest width, CW; chest depth, CD; chest circumference, CC; height across the hip, HAH; rump length, RL; hip width, HW; circumference of cannon bone, CCB. (a, b, c – P<0.05; A, B, C – P<0.01).
In this work, we recovered two indel loci in the goat ATBF1 gene for the first time, systematically explored the indel diversity of the ATBF1 gene, and analyzed their correlation with growth-related traits, resulting in the discovery of one 12-bp indel (P1-12-bp indel) and one 6-bp indel (P11-6-bp indel) in intron 3 of the ATBF1 gene associated with growth-related traits in Chinese goat breeds.
Genetic parameter analysis showed that both loci were not at Hardy–Weinberg equilibrium. Artificial selection promotes the mutation of these loci, and if these mutations only happened a few generations ago, this can be a possible explanation for the disequilibrium (Liu et al., 2017). Linkage disequilibrium analysis found that these two loci had no linkage relationship. The association analysis found that in all three breeds, the P1-12-bp indel was obviously correlated with body height and the P11-6-bp indel was obviously correlated with chest circumference and hip width. Moreover, these two loci were more or less associated with other growth-related traits. The D allele and DD genotype can be considered protogene and predominant genotypes, respectively. In SWCGs, the P1-12-bp indel also correlated with body weight (P<0.05). These results are consistent with the study on SNPs in the goat ATBF1 gene (Zhang et al., 2015), which found that polymorphisms are associated with economically important production traits.
The ATBF1 gene has a variety of functions. It plays an important roles in regulating myogenesis and adipose tissue development (Klein et al., 1996; Berry et al., 2001; Huang et al., 2015) and has a close relationship with the lactation-related hypothalamic–pituitary–adrenal (HPA) axis pathway (PITX2/PITX1–HESX1–LHX3/LHX4–PROP1–POU1F1) (Goldberg et al., 2011; Ma et al., 2017) and JAK–STAT pathways in livestock science (Nagpal et al., 2002; Ruiz-Vela et al., 2009; Ma et al., 2017). ATBF1 not only activates the expression of the Pit1 (the pituitary-specific transcription factor 1, also known as POU1F1) gene though interacting with a Pit1 enhancer (Qi et al., 2008; Araujo et al., 2013; Yan et al., 2018), but also potentially synergizes with PROP1 that can bind to the enhancer of the Pit1 gene and regulate the expression levels of the growth hormone (Li et al., 2013) prolactin and of TSH-β. STAT3, Pit1 and PROP1 genes play an important role in early embryo development and cell differentiation (Hirano et al., 2000; Ying et al., 2003; Laouar et al., 2003), so the ATBF1 gene was hypothesized to produce important effects on early development and cell differentiation; thus, it would affect the grow traits in animals.
The intronic mutation might affect the binding of the DNA sequence and DNA binding factors, such as transcription factors and splicing factors (Subaran et al., 2016; Mose et al., 2017). For example, Van-Laere et al. (2003) identified a quantitative trait locus (QTL) affecting muscle growth in pigs, which was mapped to the QTL region (IGF2-intron3-G3072A). These results indicate that regulatory mutations are important for controlling phenotypic variations. Moreover, intronic mutation might influence the transcriptional efficiency as well as the stability of mRNA (Guan et al., 2012; Minor et al., 2013; Cui et al., 2018; Wang et al., 2018).
Furthermore, association analysis of genotypes of polymorphisms and growth-related traits is an important way to evaluate the effects of a gene in animal breeding (Xu et al., 2017). These findings might provide molecular markers for MAS programs to improve the growth-related traits of local breeds in the goat industry.
Two indel loci in the ATBF1 gene were found to be significantly associated with growth-related traits in three Chinese goat breeds (P<0.05), suggesting that it can be considered an effective molecular marker for marker-assisted selection to improve economically important production traits of goat.
The original data are available upon request to the corresponding authors.
ZW and YH performed this experiment. KW wrote this paper, HY and HZ provided samples, LQ provided data, CP and XL designed this experiment, and HC checked the paper and gave advice.
The authors declare that they have no conflict of interest.
This work was supported by National Natural Science Foundation of China (no.
31760650; no. 31172184) and Provincial Key Projects of Shaanxi
(2014KTDZ02-01). We are grateful to the staff at the Shaanbei white cashmere
goat breeding farm for allowing us to collect samples and for the support
from the Agricultural Technical Station of Veterinary and Animal Husbandry of
Yulin City, Shaanxi Province, P.R. China. We are also grateful to the staff
at the Guanzhong dairy goat farm and the Hainan black goat farm for
collecting samples in Shaanxi Province and Hainan province, P.R.
China.
Edited by: Steffen Maak
Reviewed by: two anonymous referees
Aljanabi, S. M. and Martinez, I.: Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques, Nucleic Acids Res., 25, 4692–4693, 1997.
Araujo, R. V., Chang, C. V., Cescato, V. A., Fragoso, M. C., Bronstein, M. D., Mendonca, B. B., Arnhold, I. J., and Carvalho, L. R.: PROP1 overexpression in corticotrophinomas: evidence for the role of PROP1 in the maintenance of cells committed to corticotrophic differentiation, Clinics, 68, 887–891, 2013.
Berry, F. B., Miura, Y., Mihara, K., Kaspar, P., Sakata, N., Hashimoto-Tamaoki, T., and Tamaoki, T.: Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1, J. Biol. Chem., 276, 25057–25065, 2001.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, 1980.
Cui, Y., Yan, H., Wang, K., Xu, H., Zhang, X., Zhu, H., Liu, J., Qu, L., Lan, X., and Pan, C.: Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat, Frontiers in Genetics, 9, https://doi.org/10.3389/fgene.2018.00091, 2018.
Darling, D. S., Gaur, N. K., and Zhu, B.: A zinc finger homeodomain transcription factor binds specific thyroid hormone response elements, Mol. Cell. Endocrinol., 139, 25–35, 1998.
Falush, D., Stephens, M., and Pritchard, J. K.: Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies, Genetics, 164, 1567–1587, 2003.
Goldberg, L. B., Aujla, P. K., and Raetzman, L. T.: Persistent expression of activated Notch inhibits corticotrope and melanotrope differentiation and results in dysfunction of the HPA axis, Dev. Biol., 358, 23–32, 2011.
Guan, B., Gao, M., Wu, C. H., Wang, T. L., and Shih, I. M.: Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions, Neoplasia, 14, 986–993, 2012.
Hirano, T., Ishihara, K., and Hibi, M.: Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors, Oncogene, 19, 2548–2556, 2000.
Huang, Y., Wang, C., Yao, Y., Zuo, X., Chen, S., Xu, C., Zhang, H., Lu, Q., Chang, L., Wang, F., Wang, P., Zhang, R., Hu, Z., Song, Q., Yang, X., Li, C., Li, S., Zhao, Y., Yang, Q., Yin, D., Wang, X., Si, W., Li, X., Xiong, X., Wang, D., Huang, Y., Luo, C., Li, J., Wang, J., Chen, J., Wang, L., Wang, L., Han, M., Ye, J., Chen, F., Liu, J., Liu, Y., Wu, G., Yang, B., Cheng, X., Liao, Y., Wu, Y., Ke, T., Chen, Q., Tu, X., Elston, R., Rao, S., Yang, Y., Xia, Y., and Wang, P.: Molecular basis of gene-gene interaction: cyclic cross-regulation of gene expression and post-GWAS gene-gene interaction involved in atrial fibrillation, PLoS Genetics, 11, e1005393, https://doi.org/10.1371/journal.pgen.1005393, 2015.
Klein, R. D., Gu, Q., Goddard, A., and Rosenthal, A.: Selection for genes encoding secreted proteins and receptors, P. Natl. Acad. Sci. USA, 93, 7108–7113, 1996.
Laouar, Y., Welte, T., Fu, X. Y., and Flavell, R. A.: STAT3 is required for Flt3L-dependent dendritic cell differentiation, Immunity, 19, 903–912, 2003.
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., He, L., and Shi, Y.: A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn), Cell Res., 19, 519–523, 2009.
Li, M., Zhao, D., Ma, G., Zhang, B., Fu, X., Zhu, Z., Fu, L., Sun, X., and Dong, J. T.: Upregulation of ATBF1 by progesterone-PR signaling and its functional implication in mammary epithelial cells, Biochem. Bioph. Res. Co., 430, 358–363, 2013.
Liu, S., He, S., Chen, L., Li, W., Di, J., and Liu, M.: Estimates of linkage disequilibrium and effective population sizes in Chinese Merino (Xinjiang type) sheep by genome-wide SNPs, Genes Genom., 39, 733–745, 2017.
Ma, L., Qin, Q. M., Yang, Q., Zhang, M., Zhao, H. Y., Pan, C. Y., Lei, C. Z., Chen, H., and Lan, X. Y.: Associations of six SNPs of POU1F1-PROP1-PITX1-SIX3 pathway genes with growth traits in two Chinese indigenous goat breeds, Ann. Anim. Sci., 17, 399–411, 2017.
Minor, E. A., Dubovy, S., and Wang, G.: AMD-associated variants at the chromosome 10q26 locus and the stability of ARMS2 transcripts, Invest. Ophth. Vis. Sci., 54, 5913–5919, 2013.
Mose, L. E., Hayes, D. N., Perou, C. M., and Parker, J. S.: Improved indel detection in RNA-seq data via assembly based re-alignment reveals expressed Epidermal Growth Factor Receptor indels in Lung Adenocarcinoma, American Association for Cancer Research, 77, 3592, https://doi.org/10.1158/1538-7445.am2017-3592, 2017.
Nagpal, J. K., Mishra, R., and Das, B. R.: Activation of Stat-3 as one of the early events in tobacco chewing-mediated oral carcinogenesis, Cancer, 94, 2393–2400, 2002.
Nei, M.: Analysis of gene diversity in subdivided populations, P. Natl. Acad. Sci. USA, 70, 3321–3323, 1973.
Qi, Y. C., Ranish, J. A., Zhu, X. Y., Krones, A., Zhang, J., Aebersold, R., Rose, D. W., Rosenfeld, M. G., and Carriere, C.: Atbf1 is required for the Pit1 gene early activation, P. Natl. Acad. Sci. USA, 105, 2481–2486, 2008.
Ruiz-Vela, A., Aguilar-Gallardo, C., and Simón, C.: Building a framework for embryonic microenvironments and cancer stem cells, Stem Cell Rev. Rep., 5, 319–327, 2009.
Shi, Y. Y. and He, L.: SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci, Cell Res., 15, 97–98, 2005.
Subaran, R. L., Odgerel, Z., Swaminathan, R., Glatt, C. E., and Weissman, M. M.: Novel variants in ZNF34 and other brain-expressed transcription factors are shared among early-onset MDD relatives, Am. J. Med. Genet. B, 171, 333–341, 2016.
Van-Laere, A. S., Nguyen, M., Braunschweig, M., Nezer, C., Collette, C., Moreau, L., Archibald, A. A., Haley, C. S., Buys, N., Tally, M., Andersson, G., and Georges, M.: A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig, Nature, 425, 832–836, 2003.
Wang, K., Yan, H. L., Xu, H., Yang, Q., Zhang, S. H., Pan, C. Y., Chen, H., Zhu, H. J., Liu, J. W., Qu, L., and Lan X. Y.: A new indel within goat casein alpha S1 gene is significantly associated with litter size, Gene, 15, 97–98, https://doi.org/10.1016/j.gene.2018.05.119, 2018.
Wang, X. Y., Yang, Q., Wang, K., Zhang, S. H., Pan, C. Y., Chen, H., Qu, L., Yan, H. L., and Lan, X. Y.: A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats, Anim. Genet., 48, 735–736, 2017.
Xu, H., Zhang, S., Zhang, X., Dang, R., Lei, C., Chen, H., and Lan, X.: Evaluation of novel SNPs and haplotypes within the ATBF1 gene and their effects on economically important production traits in cattle, Arch. Anim. Breed., 60, 285–296, https://doi.org/10.5194/aab-60-285-2017, 2017.
Yan, H., Zhang, F., Wang, K., Liu, J., Zhu, H., Pan, C., and Qu, L.: A novel 12 bp deletion within goat LHX4 gene significantly affected litter size, Arch. Anim. Breed., 61, 1–8, https://doi.org/10.5194/aab-61-1-2018, 2018.
Yang, Q., Yan, H. L., Li, J., Xu, H., Wang, K., Zhu, H. J., Chen, H., Qu, L., and Lan, X. Y.: A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size, Anim. Genet., 48, 499–500, 2017.
Ying, Q. L., Nichols, J., Chambers, I., and Smith, A.: BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3, Cell, 115, 281–292, 2003.
Zhang, X., Wu, X., Jia, W., Pan, C., Li, X., Lei, C., Chen, H., and Lan, X.: Novel nucleotide variations, haplotypes structure and associations with growth related traits of goat at motif-binding factor (atbf1) gene, Asian Austral. J. Anim., 28, 1394–1406, 2015.
Zhao, H., Wu, X., Cai, H., Pan, C., Lei, C., Chen, H., and Lan, X.: Genetic variants and effects on milk traits of the caprine paired-like homeodomain transcription factor 2 (PITX2) gene in dairy goats, Gene, 532, 203–210, 2013.
Zhao, W., Zhong, T., Wang, L. J., Li, L., and Zhang, H. P.: Extensive female-mediated gene flow and low phylogeography among seventeen goat breeds in southwest China, Biochem. Genet., 52, 355–364, 2014.





