the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Relationships between milk protein polymorphisms and production traits in cattle: a systematic review and meta-analysis
Sinan Kopuzlu
Mehmet Topal
Omer Cevdet Bilgin
Many researchers who have studied the relationships between milk protein polymorphisms and some yield traits in dairy cattle have reported incompatible results. In this study, in order to examine the overall relationships between milk protein genes known as major genes (CSN1S1, CSN2, CSN3, and BLG) and some yield traits (daily milk yield, lactation milk yield, fat yield, fat content, protein yield, and protein content), a meta-analysis was performed using some genetic models reported in the results of previous studies on cattle. The results suggest that the relationships of major milk protein genes with other factors should be studied using the codominant genetic model in general. Relationships among some CSN3 genotypes and fat yield, fat content, and protein content, and relationships between some BLG genotypes and daily milk yield, fat content, protein yield, and protein content were significant (P<0.05). No significant (P>0.05) relationships were found between these genotypes and other milk production traits. In addition, no significant (P>0.05) relationships between the CSN1S1 and CSN2 genotypes with the milk production traits examined were observed.
- Article
(200 KB) - Full-text XML
-
Supplement
(628 KB) - BibTeX
- EndNote
For more than 50 years, many studies have reported alternative forms of milk protein genes and examined the relationships between these genetic variants and economic yield traits. Many studies look at the relationship between milk protein genetic variants and various milk production traits. Some authors reported the possibility of using milk protein as a polymorphic genetic marker (Eenennaam and Medrano, 1991; Chung et al., 1996; Kaygisiz and Dogan, 1999; Ikonen et al., 1999; Cardak, 2005; Alipanah et al., 2008; Heck et al., 2009; Mohammadi et al., 2013; Djedovic et al., 2015; Dyman et al., 2015). However, these relationships were reported to be non-significant by other researchers (Sekerden et al., 1999; Jusczak et al., 2001; Micinski et al., 2006; Ozdemir and Dogru, 2007; Gurcan, 2011; Dogru, 2015a; Molee et al., 2015).
Although numerous relationship studies between milk protein gene polymorphic systems and some economic yield traits have been published, the results are conflicting. These discrepancies may be due to differences in the sample size, breeds studied, environmental effects, gene–environment interactions, and study designs. The conflicting results of these studies, which are performed and frequently repeated in various countries, do not benefit breeders.
Meta-analysis is a test method in which the results of many studies performed in the field are used, which offers more powerful results. Importantly, the different studies used can utilize different samples and the results can be conflicting. Meta-analysis is beneficial for studies intensively repeated with an insufficient number of samples and for those in which the results are not in the proper format because sufficient criteria have been generated for the meta-analysis when the results of many studies are used. Different individual study results are combined and analyzed in the meta-analysis. Then, it is possible to achieve more powerful results in experiments using this analysis due to the randomness and sufficient sample size.
Meta-analysis has a number of potential advantages. These are an increase in power, an enhancement in accuracy, the ability to answer questions that are not responded to by individual studies, and the opportunity to resolve controversies that originate from contradictory requirements. Nevertheless, meta-analysis also has some disadvantages, one of which is the risk of serious misleading, especially when specific study designs, such as within-study biases, variation across studies, and reporting biases are not carefully analyzed. Being familiar with the type of data (e.g., categorical or continuous) which result from the measurement of an outcome in an individual study and choosing the appropriate effect measures in order to compare intervention groups bear great importance. The majority of meta-analysis methods represent variations on a weighted average of the effect estimates from various studies.
In this study, the aim was to conduct a meta-analysis on the results of previous dairy cattle studies with the purpose of analyzing the overall relationships between milk protein genes, which are known as major genes (alphas1-casein (αs1-CN, CSN1S1), beta-casein (β-CN, CSN2) kappa-casein (κ-CN, CSN3) and beta-lactoglobulin (β-LG, BLG)), and a number of production traits.
2.1 Materials and data extraction
Scientific journals were searched for published papers in English on genetic relationships between milk protein polymorphism and milk production traits in several databases (Web of Science, Science Direct, and Google Scholar), and 120 original publications from 1986 to 2016 were collected; the number of articles has been reduced for reasons such as suitability to criteria used and the presentation of results, and 40 papers were chosen. Of the collected publications, 15 consisted of alphas1-casein (10 506 yield records), 13 of them consisted of beta-casein (nearly 2223 yield records), 32 of them consisted of kappa-casein (nearly 7883 yield records), and 28 of them consisted of beta-lactoglobulin (nearly 2598 yield records). All selected papers met the three following criteria: (1) the relationship between milk protein polymorphism and milk production traits (daily milk, lactation milk yield, fat yield, fat content, protein yield, and protein content), (2) number of genotypes per animal breeds, and (3) the average and standard deviation or error of the related yield trait of each genotype (standard errors were converted to standard deviations using a statistical formula).
We extracted study content and data independently using a standard form prepared in Excel. The content of the studies included the first author's name, year of publication, country, breed, number of animals examined, number of genotypes, Hardy–Weinberg equilibrium, mean of the related yield traits of genotypes, standard deviation of means, and statistical significance level of the relationship. For the statistical analysis, this information was carefully organized by authors in order to avoid errors.
2.2 Statistical analysis
All statistical analyses were conducted using STATA version 11.2 (StataCorp 2001; Stata Statistical Software). A p value less than 0.05 was accepted as statistically significant.
The data set was separately organized for each gene region, and the methods below were followed.
-
In the meta-analysis, the analysis of differences between means is performed using a random or fixed model. Model selection was based on whether the effects of the studies were homogeneous or heterogeneous (Higgins and Thompson, 2002). The fixed model was used when the study results were homogeneous, and the random model was used when they were heterogeneous. The heterogeneity assumption was calculated based on I2 statistics (for the test of heterogeneity, the significant level (P) was set as 0.10).
-
The heritage pattern of the alleles to be used in this study was examined as dominant (AA + AB versus BB), completely overdominant (AA + BB versus AB), recessive (AA versus AB + BB), or codominant (AA versus AB, AA versus BB, and AB versus BB).
-
In the analysis of factors, genotype differences related to yield trait were evaluated separately according to the growing types of cattle breeds (dairy subgroup: dairy cattle breeds; other subgroup: dual purpose breeds, various crossbreeds; overall).
-
The standard mean differences (SMDs) and standard errors were calculated with a 95 % CI (confidence interval) to evaluate the strength of the relationship among the yield trait means of each of the gene variants examined. This procedure was used in the comparison of multiple pairwise variants.
-
In the calculation of SMD, the Cohen method (Cohen, 1988) was used when the number of studies was greater than 10, and the Hedges method (Hedges, 1981) was used when it was lower than 10. In cases in which the number of studies was > 10, the Cohen method for standardized mean differences was advantageous because it has a tendency to overestimate the effect of size. However, if the number of studies is small, the Hedges method for standardized mean differences is advantageous (DeCoster 2009; Borenstein et al., 2009).
The data sets used in this study were organized separately by economic production traits, loci, and genetic models. The dominant (AA + AB versus BB), completely overdominant (AA + BB versus AB), recessive (AA versus AB + BB), and codominant (AA versus AB, AA versus BB, and AB versus BB) statuses of the alleles were taken into consideration before the statistical evaluation (Table 1). According to the analysis results, evaluations were mainly carried out based on this model in the paper because the relevant alleles generally exhibited the codominant trait.
The number of studies and the meta-analysis results in which different methods (Cohen or Hedges) were used based on whether the studies were homogeneous or heterogeneous are presented in Tables 1, 2, 3, 4, and 5. The heterogeneity test, SMD, 95 % CI values, its % weight, and P values of the pairwise comparisons are presented in the tables as well.
Table 1Genetic model analyses of the genotypes related to BLG, CSN2, and CSN3.
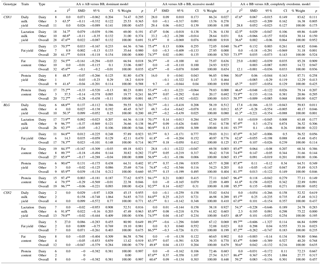
* p<0.10. P<0.01. n number of publication.
3.1 Analysis of CSN3 genotypes
Using 13 studies, no significant relationships could be determined between CSN3 genotypes (AA vs. AB, AA vs. BB, and AB vs. BB) and daily milk yield in all cattle and subgroups (P>0.05). In terms of daily milk yield, although the ranking was observed to be AA > AB > BB in the dairy group, it was observed to be exactly the opposite in the other group. However, when analyses were performed according to the recessive model, the dairy subgroup AA vs. AB + BB genotype mean difference was found to be statistically significant (P<0.05) and was observed to provide advantages in favor of AA (Table 1). In some individual studies performed previously, it was reported that CSN3 AA and AB genotypes had superiority (Cardak, 2005; Pawelska-Goral et al., 2008; Bartonova et al., 2012; Molee et al., 2015). Some researchers reported that daily milk yield means were not significant among CSN3 genotypes (Dogru, 1994; Ozdemir and Dogru, 2005; Gurcan, 2011).
Table 2Results of the meta-analysis on associations among CSN3 genotypes and milk production traits, SMD values, and level of significance.
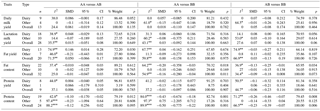
* p<0.10. P<0.01. n number of publication.
When 28 studies were used, a significant relationship could not be determined between CSN3 genotypes and lactation milk yield in all cattle and milk type cattle groups (P>0.05). In the analysis performed according to the recessive genetic model, a significant difference was observed in favor of the AA-only genotype in the other subgroup (P<0.05); significant differences among groups were not observed when compared to other models (P>0.05). In the individual studies performed, some authors reported that lactation milk yield means were significant among CSN3 genotypes (Dogan and Kaygisiz, 1999; Alipanah et al., 2008; Djedovic et al., 2015), and other authors reported that mean differences were not significant (Eenennaam and Medrano, 1991; Bovenhuis et al., 1992; Kaygisiz et al., 1999; Ozdemir and Dogru, 2005; Dogru, 2015a).
Using 20 studies, a significant relationship could not be established between CSN3 genotypes and fat yield in all cattle (P>0.05); however, the AA vs. AB genotype fat yield means were found statistically different (P<0.05) and the AA genotype was found superior in the dairy subgroup. In the other subgroup, statistically significant relationships were not determined between fat yield means belonging to genotypes (P>0.05). According to the completely overdominant model, statistical significance was determined against AB in the dairy subgroup and in favor of the AB variant in the other subgroup (P<0.05). In the analysis performed according to the recessive model, while a significance in favor of the AA genotype was determined in dairy subgroup, a significant difference for only the AA genotype was found in the other subgroup (P<0.05). In the individual studies performed, while some authors reported that fat yield means were significant among the CSN3 genotypes and reported a superiority in favor of the AB genotype (Alipanah et al., 2008; Dogru, 2015a; Djedovic et al., 2015), other authors reported that the mean differences were not significant (Eenennaam et al., 1990; Bovenhuis et al., 1992; Cardak, 2005; Ozdemir and Dogru, 2005; Zhang et al., 2007; Heck et al., 2009; Alim et al., 2015). Consequently, as a result of the meta-analysis performed, the use of the CSN3 AB variant in programs that seek to improve fat yield will be useful as an indirect marker.
A total of 32 studies were used to assess the relationship between CSN3 genotypes and fat content. A statistically significant relationship (P<0.05) was determined between AA vs. BB genotypes in all cattle and fat content and between AA vs. BB and AB vs. BB genotypes in the dairy subgroup. Significant differences could not be found in terms of fat content in the other groups examined or among the other genotypes. Among all the models analyzed, only the dominant model revealed that the BB genotype in all the cattle and the dairy subgroup was different.
According to our results, there is a selection advantage in favor of BB genotype for milk fat content, and it can be used as a marker gene. In the individual studies performed, some authors reported that the fat content means were significantly different among the CSN3 genotypes and reported a superiority in favor of the BB genotype (Bovenhuis et al., 1992; Alipanah et al., 2008; Dogru, 2015a); other authors reported that the mean differences were not significant (Eenennaam and Medrano, 1991; Cardak, 2005; Ozdemir and Dogru, 2005; Zhang et al., 2007; Heck et al., 2009; Djedovic et al., 2015).
The mean differences of the genotype groups examined for protein yield were found not significant according to all the genetic models (P>0.05). In individual studies performed, some authors reported differences between protein yield means and different CSN3 genotypes, including a significant preference for the BB genotype (Eenennaam and Medrano, 1991; Alim et al., 2015); however, other authors did not find these differences significant (Bovenhuis et al., 1992; Micinski et al., 2006; Alipanah et al., 2008).
In the analysis in which 24 studies were used for assessing the relationship between CSN3 genotypes and protein content, a statistically high significant relationship (P<0.01) was determined between AA vs. BB and AB vs. BB genotypes in all cattle in terms of protein content. The relationship between AA vs. AB was found not significant (P>0.05) and the genotypic ranking in terms of protein content was found to be BB > AB > AA. A statistically significant relationship was determined between the protein content values and all CSN3 genotypes in the dairy subgroup, and the ranking of genotypes was observed to be BB > AB > AA (Table 1). A significant relationship could not be determined between the protein content means and any of the genotypes in the other subgroup (P>0.05). The BB variant was seen as advantageous when the analysis was performed according to the dominant model, and a significant relationship against the AA variant was found when the recessive model was used (Table 1). Based on the results for other genetic models, the use of the CSN3 B allele as a marker to improve milk protein content in dairy cattle breeds should be considered.
3.2 Analysis of BLG genotypes
A total of 10 studies were used to assess the relationship between BLG genotype (AA vs. AB, AA vs. BB, and AB vs. BB) and daily milk yield, and highly significant differences were observed in all cattle and subgroups (P<0.01). Although only the difference among the AB vs. BB genotypes was found significant in the dairy subgroup (P<0.01), the differences between the means of AA vs. AB (P<0.01) and AA vs. BB (P<0.05) genotypes were found to be significant in the other subgroup. Although the differences between AA vs. AB (P<0.05) and AB vs. BB (P<0.01) genotype means were significant in all cattle, a difference among AA vs. BB means was not significant (P>0.05). In terms of daily milk yield, AB > BB > AA. When using the completely overdominant model, the mean was found to be statistically superior in favor of AB. In individual studies performed, some researchers reported that the daily milk yield means were significant among BLG genotypes (Cardak, 2005; Molee et al., 2015); however, other researchers reported that the mean differences were not significant (Sekerden et al., 1999; Ozdemir and Dogru, 2007).
Table 3Results of the meta-analysis on the associations among BLG genotypes and milk production traits, SMD values, and level of significance.
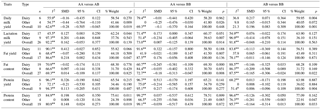
* p<0.10. P<0.01. n number of publication.
Table 4Results of the meta-analysis on associations among CSN2 genotypes and milk production traits, SMD values, and level of significance.
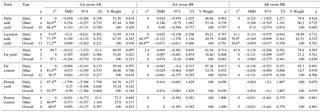
* p<0.10. P<0.01. n number of publication.
Table 5Results of the meta-analysis on associations among CSN1S1 genotypes and milk production traits, SMD values, and level of significance.
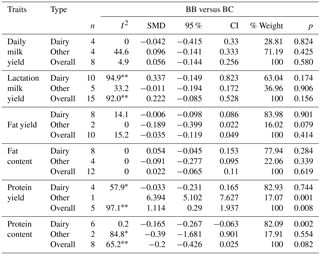
* p<0.10. P<0.01. n number of publication.
In the analysis in which 26 studies were used, a significant relationship was not observed between BLG genotypes and lactation milk yield in all cattle (P>0.05); however, a statistically significant relationship (P<0.05) was observed between AA vs. AB genotypes in the milk type group and between AA vs. BB genotypes in the other subgroup in terms of milk yield means. Superiority was observed in favor of AA for the dairy subgroup and in favor of BB for the other subgroup. The differences among lactation milk yield means for the other genotype pairs examined were not significant (P>0.05). Similar results were observed in the analyses performed according to the other genetic models. In the individual studies performed, some researchers reported that the differences in lactation milk yield means among BLG genotypes were significant (Kaygisiz et al., 1999; Ojala et al., 2004; Heidari et al., 2009), yet other researchers reported that mean differences were not significant (Eenennaam and Medrano, 1991; Ozdemir and Dogru, 2007; Dokso et al., 2011; Dogru, 2015b).
In the analysis in which 17 studies were used, a significant relationship was not observed between BLG genotypes and fat yield in the subgroups (P>0.05). Although a statistically significant relationship (P<0.05) was determined among AA vs. AB genotype means in all cattle, significant differences were not found among the other genotype means. The differences among the genotype means compared were not found significant in the analyses performed according to other genetic models. In the individual studies performed, some researchers reported the differences between fat yield means and BLG genotypes as significant and in favor of the BB genotype (Ojala et al., 2004; Felenczak et al., 2008), yet other researchers could not find significant differences (Eenennaam and Medrano, 1991; Ozdemir and Dogru, 2007; Micinski et al., 2006; Czerniawska et al., 2011; Dogru, 2015b).
A total of 27 studies were used to assess the relationship between the BLG genotypes and fat content. Although a statistically significant relationship (P<0.01) was found between AA vs. BB (P<0.01) and AB vs. BB (P<0.05) genotypes in all cattle in terms of fat content and among AA vs. BB genotypes in the dairy subgroup, significant differences were not found in terms of fat content between the genotypes in the other subgroup (P>0.05). When the analysis was performed according to the dominant model, the fat content values of the BB variant were higher, and the difference was statistically significant. Using the recessive model, the AB + BB variant in the dairy subgroup was also statistically superior when compared to the fat content value of the AA variant (Table 1). These results suggest that the BLG B allele can be used as a genetic marker in programs that seek to alter fat content. In the individual studies performed, some researchers reported that the BLG BB variant was significantly advantageous in terms of fat content values (Juszczak et al., 2001; Ojala et al., 2004; Czerniawska et al., 2011; Molee et al., 2015). However, other studies reported that the differences among the fat content values of the different genotypes were not significant (Eenennaam and Medrano, 1991; Felenczak et al., 2008; Micinski et al., 2006; Ozdemir and Dogru, 2007; Dokso et al., 2011; Dogru, 2015b).
In the groups examined, while BLG was seen as AA > AB > BB among the protein yield means of all genotypes, differences among mean values were not statistically significant (P>0.05). The differences among the protein yield means of BLG genotypes compared were not found significant in the analyses performed according to other genetic models (P>0.05). In individual studies performed, while the differences among protein yield means of BLG genotypes were reported to be significant in some studies (Ojala et al., 2004; Felenczak et al., 2008), these differences were not significant in other studies (Eenennaam and Medrano, 1991; Micinski et al., 2006; Czerniawska et al., 2011).
A total of 19 studies were used to examine the relationship among the BLG genotype and protein content. Although a statistically significant relationship (P<0.05) was found between AA vs. AB and AB vs. BB genotypes in all cattle in terms of protein content, the relationship among AA vs. BB genotypes was not significant (P>0.05). While a statistically significant relationship (P<0.05) was observed among the AA vs. AB genotypes in the dairy subgroup and among the AB vs. BB genotype protein content values in the other subgroup, significant differences were not found among protein content values for the other genotypes examined (P>0.05). Based on these results, we conclude that the BLG homozygote genotypes are superior and this should be taken into consideration during selective breeding. In other genetic model analyses, the AA genotype was found to be superior in the dairy subgroup using the recessive model; superiority in favor of the BB genotype was just observed in the other subgroup using the dominant model. No significant relationships were found between the protein content values and other genotypes. In the individual studies performed, while some authors found the differences among protein content means in the various BLG genotypes significant (Ojala et al., 2004; Felenczak et al., 2008; Mohammadi et al., 2013; Dyman et al., 2015), other authors did not find the differences significant (Eenennaam and Medrano, 1991; Ikonen et al., 1999; Botara et al., 2009; Molee et al., 2015).
3.3 Analysis CSN2 genotypes
Few publications have examined CSN2. In the analyses to assess the relationship between the genotypes and yield traits, fat yield and AA vs. AB genotypes were found statistically significant only in the other subgroup (P<0.05); no significant relationships were observed for any of the other genotypes and traits (P>0.05). In the analyses performed using other genetic models, only the fat yield values for genotypes in the other subgroup were found significant when using the recessive and completely overdominant models. The difference in genotype mean values was significant in the other subgroup for lactation milk yield also. The differences among all other genotype means were not significant (P>0.05). In some of the individual studies examining CSN2, differences among the CSN2 genotypes were reported to be significant regarding daily milk yield (Heck et al., 2009; Gurcan, 2011; Molee et al., 2015), lactation milk yield (Dogru, 1994; Ikonen et al., 2001; Micinski et al., 2006), protein content (Cardak, 2005; Micinski et al., 2006), fat content (Dogru, 1994; Cardak, 2005), and fat yield (Dogru, 1994). In other studies, these differences in genotypes were not significant for daily milk yield (Dogru, 1994; Gurcan, 2011; Ozdemir and Dogru, 2007), lactation milk yield (Eenennaam and Medrano, 1991; Kaygisiz et al., 1999; Ozdemir and Dogru, 2007), protein content and fat content (Eenennaam and Medrano, 1991; Sekerden et al., 1997; Micinski et al., 2006), and protein yield and fat yield (Eenennaam and Medrano, 1991; Sekerden et al., 1997; Micinski et al., 2006). A study conducted by Ikonen et al. (2001) indicated that CSN2 BB variants might be useful in direct selection programs for improving milk and fat yield. This finding is further supported by a study conducted by Chung et at. (1996) that reported a highly significant relationship between the CSN2 AA variant and fat content. However, our meta-analysis results indicate that none of the CSN2 variants provide an advantage, indicating that they are not particularly useful for improvement programs.
3.4 Analysis of CSN1S1 genotypes
Few CSN1S1 studies were suitable for the meta-analyses, so only the relationship between the BB and BC genotypes and yield traits could be examined. Previous studies did not provide sufficient data to assess the other genotypes. According to the results of the meta-analysis performed, highly significant differences (P<0.01) were seen between the BB and BC genotype values in terms of protein content in the dairy subgroup and in terms of protein yield in the other subgroup and in all cattle. The differences for other yield traits were not significantly different among other genotypes (P>0.05). In the individual studies performed, significant differences among genotypes in terms of fat content, fat yield, protein content, and protein yield were found by a number of authors (Bovenhuis et al., 1992; Cardak, 2005); in contrast, many researchers reported the relationships between the yield traits and CSN1S1 genotypes as not significant (Bovenhuis et al., 1992; Ozdemir and Dogru, 2004; Cardak, 2005; Micinski et al., 2006; Gurcan, 2011; Hristov et al., 2013). Our meta-analysis results indicate that CSN1S1 variants will not provide any advantage in improvement programs for any of the traits examined.
In this meta-analysis, the relationships between major milk protein genes and some economic yield traits were analyzed and evaluated according to several genetic models. The meta-analysis demonstrated that the relationships of major milk protein genes with other factors should be examined using the codominant genetic model in general. Significant relationships were determined for the CSN3 genotypes and fat content and protein content when examined using the meta-analysis. Significant relationships were also determined among the BLG genotypes and daily milk yield, fat content, protein yield, and protein content. This suggests that these major genes will be useful for the improvement of the economic yield traits examined and can be used as molecular markers. Furthermore, our analyses generally showed that there were not relationships between the CSN1S1 and CSN2 polymorphisms and any of the yield traits examined, and these genes will not be useful as markers for the improvement of traits. Moreover, the meta-analysis performed helped to clarify contradictory studies present in the field.
Data sets are not deposited in different repositories and data from a third party were not used. the data is original and users can connect with your site by link.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-61-197-2018-supplement.
The authors declare that they have no conflict of
interest.
Edited by: Steffen Maak
Reviewed by: Cigdem Takma and Eser Gurcan
Alim, M. A., Sun, D., Zhang, Y., Zhang, Y., Zhang, Q., and Liu, L.: DNA Polymorphisms in the β-lactoglobulin and κ-casein Genes Associated with Milk Production Traits in Dairy Cattle, Biores. Comm., 1, 82–86, 2015).
Alipanah, M. L., Alexandrovna, K. G., and Veladimirovich, R.: Kappa-casein and prl-rsa i genotypic frequencies in two russian cattle breeds, Arch. Zootec., 57, 131–138, 2008.
Bartonova, P., Vrtkova, I., Kaplanova, K., and Urban, T.: Association between CSN3 and BCO2 gene polymorphisms and milk performance traits in the Czech Fleckvieh cattle breed, Genet. Mol. Res., 11, 1058–1063, 2012.
Borenstein, M., Hedges, L. V., Higgins, J. P. T., and Rothstein, H. R.: Introduction to Meta-Analysis, John Wiley & Sons, United Kingdom, p. 402, 2009.
Botaro, B. G., Lima, Y. V. R., Aquino, A. A., Fernandes, R. H. R., Garcia, J. F., and Santos, M. V.: Effect of beta-lactoglobulin polymorphism and seasonality on bovine milk composition, J. Dairy Res., 75, 176-181, https://doi.org/10.1017/S0022029908003269, 2008.
Bovenhuis, H., Arendonk, J. A. M., and Korver, S.: Associations between milk protein polymorphisms and milk production traits, J. Dairy Sci., 75, 2549–2559, https://doi.org/10.3168/jds.S0022-0302(92)78017-5, 1992.
Burbano, G. L. Z., Cabrera, Y. M. E., Portilla, C. E. S., and Galindo, C. Y. R.: Kappa casein genotypes and curd yield in Holstein cows, Rev. Colomb. Cienc. Pecu., 23, 422–428, 2010.
Cardak, A. D.: Effects of genetic variants in milk protein on yield and composition of milk from Holstein-Friesian and Simmentale cows, S. Afr. J. Anim. Sci., 35, 41–47, https://www.ajol.info/index.php/sajas/article/viewFile/4047/11890, 2005.
Celik, S.: Beta-lactoglobulin genetic variants in Brown Swiss breed and its association with compositional properties and rennet clotting time of milk, Intern. Dairy J., 13, 727–731, 2003.
Chung, H. Y., Lee, H. K., Chen, K. J., You, C. H., Park, K. D., Kim, K. N., and Chung, E. R.: Studies on the relationships between biochemical polymorphisms and production traits in dairy cattle, Korean J. Dairy Sci., 18, 7–16, 1996.
Cohen, J.: Statistical Power Analysis for the Behavioral Sciences 2 Edn., Hillsdale, NJ, Erlbaum, 1988.
Czerniawska, P. E., Szewczuk, M., Olszewska, A., and Chocilowicz, E.: Association between beta-lactoglobulin (lgb) polymorphismand yield and composition of milk of holstein-friesian cows imported from Sweden, Acta Sci. Pol. Zootechnica, 10, 9–18, 2011.
Deb, R., Singh, U., Kumar, S., Singh, R., Sengar, G., and Sharma, A.: Genetic polymorphism and association of kappa-casein gene with milk production traits among Frieswal (HF × Sahiwal) cross breed of Indian origin, IJVR, 15, 406–408, 2014.
DeCoster, J.: Meta-Analysis Notes, Institute for Social Science Research, University of Alabama, available at: http://www.stat-help.com/notes.html (last access: 15 January 2016), 2009.
Djedovic, R., Bogdanovic, V., Perisic, P., Stanojevic, D., Popovic, J., and Brka, M.: Relationship between genetic polymorphism of k-casein and quantitative milk yield traits in cattle breeds and crossbreds in Serbia, Genetika, 47, 23–32, 2015.
Dogan, M. and Kaygisiz, A.: Relationships Between Milk Yield Traits and Milk Protein Polymorphism in Brown Swiss Cattle in Turkey, Tr. J. Vet. Anim. Sci., 23, 47–49, 1999.
Dogru, U.: Genetic structure of several polymorphic blood and milk proteins of Brown Swiss, Holstein, Simmental and Eastern Anotolian Red cattle breeds and their diffrences according to several production traits, PhD Thesis, Ataturk University, Erzurum, Turkey, 1994.
Dogru, U.: Kappa casein genetic variants in Holstein dairy cattle and their association with yield and quality of milk, Res. J. Anim. Sci., 9, 4–7, 2015a.
Dogru, U.: β-lactoglobulin genetic variants in Brown-Swiss dairy cattle and their association with milk yield and quality traits, J. Anim. Plant. Sci., 25, 595–598, 2015b.
Dokso, A., Kelava, N., Brka, M., and Ivankovic, A.: The effect of lgb genes on quantitative and qualitative characteristics of milk Holstein breed in Croatia. Proceedings 22nd International Scientific-Expert Conference of Agriculture and Food Industry-Sarajevo, 2011.
Dyman, T. M. and Plivachuk, O. P.: Effect of Beta-lactoglobulin genotypes on composition and technological properties of milk in Ukrainian Black and White dairy cattle, 49, 187–192, available at: http://nbuv.gov.ua/UJRN/rgt (last access: 10 February 2016), 2015.
Eenennaam, A. V. and Medrano, J. F.: Milk protein polymorphisms in California dairy cattle, J. Dairy. Sci., 74, 1730–1742, 1991.
Felenczak, A., Fertig, A., Gardzina, E., Ormian, M., and Trela, J.: Technological traits of milk of Simmental cows as related to κ-casein polymorphism, Ann. Anim. Sci., 6, 37–43, 2006.
Felenczak, A., Gil, Z., Adamczyk, K., Zapletal, P., and Frelich, J.: Polymorphism of milk κ-casein with regard to milk yield and reproductive traits of Simmental cows, J. Agrob., 25, 201–207, 2008a.
Felenczak, A., Gil, Z., Jezovit-Jurek, M., Gil, Z., Adamczyk, K., Zapletal, P., and Frelich, J.: Polymorphism of milk β-lactoglobulin and effect on milk casein with regard to milk yield and reproductive traits of Simmental cows, Ann. Anim. Sci., 8, 207–213, 2008b.
Gurcan, E. K.: Association between milk protein polymorphism and milk production traits in black and white dairy cattle in Turkey, Afr. J. Biotech., 10, 1044–1048, 2011.
Hamza, A. E., Wang, X. L., and Yang, Z. P.: Kappa casein gene polymorphism in Holstein Chinese cattle, Pak. Vet. J., 30, 203–206, 2010.
Heck, J. M. L., Schennink, A., Valenberg, H. J. F., Bovenhuis, H., Visker, M. H., Arendonk, J. A. M., and Hooijdonk, A. C. M.: Effects of milk protein variants on the protein composition of bovine milk, J. Dairy. Sci., 92, 1192–1202, 2009.
Hedges, L. V.: Distribution theory for Glass's estimator of effect size and related estimators, Journal of Educational Statistics, 6, 107–128, 1981.
Heidari, M., Azari, M. A., Hasani, S., Khanahmadi, A., and Zerehdaran, S.: Association of genetic variants of β-lactoglobulin gene with milk production in a herd and a superior family of Holstein cattle, Iranian J. Biotech., 7, 254–257, 2009.
Higgins, J. P. T. and Thompson, S. G.: Quantifying heterogeneity in a meta-analysis, Statistics in Medicine, 21, 1539–1558, 2002.
Hristov, P., Teofanova, D., Mehandzhiyski, I., Zagorchev, L., and Radoslavov, G.: Significance of Milk protein genes polymorphsm for Bulgarian Rhodopean cattle: comparative studies, Biotechnology and Biotechnological Equipment, 27, 3659–3664, 2013.
Ikonen, T., Ojala, M., and Ruottinen, O.: Associations between milk protein polymorphism and first lactation milk production traits in Finnish Ayrshire cows, J. Dairy. Sci., 82, 1026–1033, 1999.
Juszczak, J., Erhardt, G., Kuczaj, M., Zieminski, R., and Panicke, L.: Zusammenhang zwischen κ-casein und β-Lactoglobulin Varianten mit der Milchleistung und der Nutzungsdauer von Rindern der Schwarzbuntes Rind und Polnisches Rotvieh, Archiv Tierzucht Dumm, 44, 239–249, 2001.
Kaygisiz, A. and Doqan, M.: Genetics of milk protein polymorphism and its relation to milk yield traits in Holstein cows, Tr. J. Vet. Anim. Sci., 23, 447–454, 1999.
Kaygisiz, A.: Possibilities Milk Protein Polymorphism For Genetic Improvements of Simmental Cattle's. Kahramanmaras Sutcu Imam University, Faculty of Agriculture, BAP, No:98.8.18, 2000.
Lin, C. Y., McAllister, A. J., Ng-Kwai-Hang, K. F., and Hayes, J. F.: Effects of milk protein loci on first lactation production in dairy cattle, J. Dairy. Sci., 69, 704–712, 1986.
Lukac, D., Jovanovac, S., Nemes, Z., Vidovic, V., Popovic-Vranjes, A., Raguz, N., and Lopicic-Vasic, T.: Association of polymorphism κ-casein gene with longevity and lifetime production of Holstein-Friesian cows in Vojvodina, Mljekarstvo 65, 232–237, 2015.
Micinski, J. and Klupczynski, J.: Correlations between polymorphic variants of milk proteins, and milk yield and chemical composition in black-and-white and jersey cows, Pol. J. Food. Nutr. Sci., 15, 137–143, 2006.
Mir, S. N., Ullah, O., and Sheikh, R.: Genetic polymorphism of milk protein variants and their association studies with milk yield in Sahiwal cattle, Afr. J. Biotechnol., 13, 555–565, https://doi.org/10.5897/AJB2013.13216, 2014.
Mohammadi, Y., Aslaminejad, A. A., Nassiri, M. R., and Koshkoieh, A. E.: Allelic polymorphism of K-casein, β-Lactoglobulin and leptin genes and their association with milk production traits in Iranian Holstein cattle, J. Cell. Mol. Res., 5, 75–80, 2013.
Molee, A., Mernkrathoke, P., and Poompramun, C.: Effect of casein genes-beta-LGB, DGAT1. GH, and LHR-on milk production and milk composition traits in crossbred Holsteins, Genetics and Molecular Research, 14, 2561–2571, 2015.
Ojala, M., Seppanen, T., Tyriseva, A. M., and Ikonen, T.: Effects of milk protein genotypes on body weight and milk production traits in Finnish Ayrshire cows. X Baltic Animal Breeding Conference, Tartu, Estonia, 13–14 May 2004, Proc Animal Breeding in the Baltics, 68–73, 2004.
Ozdemir, M. and Dogru, U.: Relationships between Alfa-casein polymorphism and production traits in Brown Swiss and Holstein, J. Appl. Anim. Res., 25, 5–8, 2004.
Ozdemir, M. and Dogru, U.: Relationships between Beta-Lactoglobulin and Beta-casein polymorphisms and production traits in Brown Swiss and Holstein, J. Appl. Anim. Res., 31, 165–168, 2007.
Ozdemir, M. and Dogru, U.: Relationships between Kappa-casein polymorphism and production traits in Brown Swiss and Holstein, J. Appl. Anim. Res., 27, 101–104, 2005.
Pawelska-Goral, M., Zygadlik, K., and Szulc, T.: Effect of protected methionine and fishmeal addition on the composition and physical properties of milk from cows with different kappa-casein genotypes, Ann. Anim. Sci., 8, 233–241, 2008.
Sekerden, O., Dogrul, F., and Erdem, H.: Blood and milk protein polymorphism with and their effect on various production traits inseveral production traits for Simmental cows, Tr. Vet. Anim. Sci., 23, 87–93, 1999.
Trakovicka, A., Moravcíkova, N., and Navratilova, A.: Kappa-casein gene polymorphism (CSN3) and its effect on milk production traits, Acta fytotechnica et zootechnica, Nitra, Slovaca Universitas Agriculturae Nitriae 3, 61–64, 2012.
Vidovic, V., Lukac, D., Nemes, Z., and Trivunovic, S.: β-lactoglobulin genetic variants in Serbian Holstein-Friesian dairy cattle and their association with yield and quality of milk, Animal Science Papers and Reports, 32, 179–182, 2014.
Zhang, J., Sun, D., Womack, J. E., Zhang, Y., Wang, Y., and Zhang, Y.: Polymorphism Identification, RH Mapping and Association of α-Lactalbumin Gene with Milk Performance Traits in Chinese Holstein, Asian-Aust. J. Anim. Sci., 20, 1327, 2007.





