the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
An examination of melanogenic traits and TYRP1 polymorphism in Nanping and Romney Marsh sheep breeds
Gouzhi Li
Heli Xiong
Dongmei Xi
Sameeullah Memon
Liping Wang
Xiangying Liu
Weidong Deng
The effects of mutations of the gene for tyrosinase-related protein 1 (TYRP1) on the black muscles and coat color in Nanping black-boned sheep were investigated. Tyrosinase activity and melanin content in plasma were measured and compared in three random groups of sheep: Nanping black-boned (101 heads), Nanping normal (106 heads) and Romney Marsh sheep (82 heads, Ovis aries). Eight exons and their partial flanking regions of the TYRP1 gene were amplified. Six intronic mutations and six exonic polymorphisms including two non-synonymous mutations [c.203C > T (p.A68V) and c.1202T > C (p.V401A)] were identified. Using a bi-directional polymerase chain reaction allele-specific amplification (bi-PASA) of the mutation c.203C > T it was shown that the frequencies of allele C in the Nanping black-boned, Nanping normal and Romney Marsh sheep were respectively 0.955, 0.967 and 0.744. For the mutation c.1202T > C, the frequencies of allele T in the three populations of sheep were respectively 0.777, 0.745 and 0.793 as measured using the single-strand conformation polymorphism. When the data from sheep of all three populations with the CC genotype of SNP c.203C > T were pooled, it was found that there was significantly higher (P < 0.05) tyrosinase activity, content of alkali-soluble melanin and ratio of eumelanin : total melanin than in the plasma of sheep with the CT and TT genotypes. This was not so within each of the three groups of sheep. No significant effect of the TRYP1 genotype on coat color was found. Further studies will be necessary to determine the cause of the black traits in Nanping black-boned sheep.
- Article
(3408 KB) - Full-text XML
- BibTeX
- EndNote
The black-brown pigment eumelanin and a related reddish-yellow pigment, phaeomelanin, are found naturally in the tissues of both invertebrates and vertebrates (Ito and Wakamatsu, 2008; Dubey and Roulin, 2008). The integument color in mammals is mostly determined by the ratio and quantities of these two pigments. These pigments are synthesized in cutaneous melanocytes and transported to surrounding epidermal keratinocytes (Vachtenheim and Borovanský, 2010; de Araújo Lima et al., 2015). L-tyrosine to dopamine conversion in the synthesis of melanin within melanocytes is catalyzed by the tyrosinase (EC1.14.18.1) (Hall and Orlow, 2005). Other factors and enzymes also regulate the amount and quality of the melanin synthesized (Bellei et al., 2011; Aydin et al., 2012). Furthermore, sheep melanocytes (melanogenesis) will spontaneously lose their partial function during a period of weeks to months after birth (Aliev et al., 1987).
Tyrosinase-related protein 1 (TYRP1), encoded by the TYRP1 gene, catalyzes the oxidation of 5,6dihydroxyindole-2-carboxylic acid (DHICA) to indole-5,6quinone-2-carboxylic acid at a point within the pathway for the synthesis of eumelanin (Toyoda et al., 2004). The activity of tyrosinase is increased by TYRP1 stabilizing its structure (Manga et al., 2000; Slominski et al., 2004). The TYRP1 gene has been cloned and there are numerous reports on the effects of mutations of the gene on skin and coat color in mammals, including humans (Sturm et al., 1995; Rooryck et al., 2006), mice (Zdarsky et al., 1990), goats (Becker et al., 2015), sheep (Gratten et al., 2007, 2008), cattle (Berryere et al., 2003; Guibert et al., 2004), pigs (Okumura et al., 2005, 2008), horses (Rieder et al., 2001), cats (Schmidt-Kuntzel et al., 2005; Lyons et al., 2005), dogs (Schmutz et al., 2002) and rabbits (Utzeri et al., 2014) as well as non-mammalian species including chickens (April et al., 1998) and quail (Nadeau et al., 2007; Xu et al., 2013). Moreover, it has been highly expressed in black skin (Fan et al., 2013).
We have reported the identification of black-boned sheep (Ovis aries) within a population of Nanping indigenous sheep. These black-boned sheep have dark-colored (black) tissues compared to the reddish coloration of the tissues of the normal sheep in the flock (Deng et al., 2006). The trait for the dark coloration has been shown to be inherited in cross-breeding studies (Deng et al., 2008) and the Nanping black-boned sheep have been recognized as a novel genetic resource by the Chinese Ministry of Agriculture.
In the present study we cloned the eight exons of the ovine TYRP1 gene with a view to identifying potential polymorphisms and to determining whether polymorphisms of the gene might explain the black traits and coat color of the Nanping black-boned sheep.
All experiments performed in this study were approved by the International Animal Care and Use Committee of the Yunnan Agricultural University. The care and use of animals fully complied with local animal welfare laws, guidelines and policies.
Three groups of sheep (Ovis aries) were used for the study. Two groups of sheep more than 1 year of age were identified in a flock of indigenous sheep in Nanping County, Yunnan Province, PRC. A group of 101 were classified as Nanping black-boned (NPBB) sheep and a second group of 106 were classified as Nanping normal (NPN) sheep. A third group of 82 mature Romney Marsh (RM) sheep were kept at the Yunnan Malong Sheep Stud Farm, Malong County, Yunnan Province, PRC. All sheep grazed on pasture consisting of natural grasses and herbs during the day and were penned at night.
A blood sample was collected from each sheep in the morning prior to releasing the sheep to pasture. The blood was collected from the external jugular vein by direct venipuncture, and each sample was distributed to six aliquots into sterile tubes. Sodium citrate was added to six of the aliquots prior to centrifugation to obtain plasma. All three tubes of blood or plasma were then snap frozen in liquid nitrogen before storing at −80∘ C pending analyses.
At the time of collection of blood the wool color of each sheep was recorded as either black, white mixed (black, brown and white) or brown.
Whole blood samples from five sheep chosen randomly from each of the NPBB, NPN and RM groups were used for the identification of polymorphisms. Genomic DNA was extracted from each blood sample using a TIA Namp Blood Kit (Tiangen Biotech Co. Ltd., Beijing, China). The reference genomic sequence was obtained from the sheep (EF102110) TYRP1 gene complete coding region, which has been posted on COW BLAT Search (http://genome.ucsc.edu/cgi-bin/hgBlat).
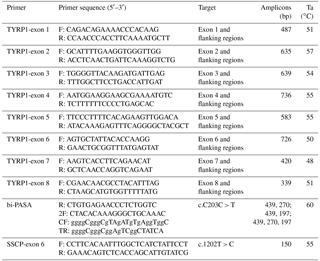
Primers were designed in inferential introns using primer Premium 5.0 software to provide optimal polymerase chain reaction (PCR) products. Eight primers were designed to amplify each exon and its partial flanking region of the TYRP1 gene (Table 1). The PCR mixture contained 2 µL (20–50 ng) of genomic DNA, 1.5 µL of 25 mM Mg Cl2, 1.0 µL of 10 pmol µL−1 forward primer, 1.0 µL of 10 pmol µL−1 reverse primer, 0.5 µL of 10 pmol µL−1 dNTP (deoxy-ribonucleoside triphosphate) mixture, 0.4 µL of 5 U Taq polymerase and 2.5 µL of 10 × PCR buffer at a final volume of 25 µL. Each PCR reaction was performed in a Gene Thermal Cycle (Hangzou, China) using the protocol shown in Table 1. The PCR program started with denaturation at 94 ∘C for 3 min, followed by 38 cycles of 94 ∘C for 45 s, Ta (annealing temperature) for 45 s, 72 ∘C for 60 s then an extended 72 ∘C cycle for 5 min and finally 4 ∘C to terminate the reaction. The PCR products were sequenced bi-directionally using an ABI 3730 DNA analyzer (Applied Biosystems Inc.) at the Sun Biotechnology Company (Beijing, China).
All collected samples except those sequenced previously were genotyped as follows.
Due to the lack of a suitable restriction nuclease for the single nucleotide polymorphism (SNP) g.203C > T, a bi-directional PCR allele-specific amplification (bi-PASA) system was designed to quickly and conveniently detect the genotype of SNP g.203C > T. The Bi-PASA method, based on a one-tube reaction, permits a distinction to be made between the different genotypes by using two outside primers and two inner (allele-specific) primers at the mutation point (Liu et al., 1997).
The bi-PASA PCR reactions were performed in a mixture (25 µL) containing 2 µL of genomic DNA, 1 µL of 25 mM Mg Cl2, 0.25 µL of 10 mM outside primer, 0.5 µL of 10 mM inner primer (Table 1), 0.5 µL of 20 mM dNTP mixture, 0.2 µL of 5 U Taq polymerase, 3 µL of 10 × buffer and 16.8 µL of water. The thermal cycling PCR conditions were 94 ∘C for 3 min, 40 cycles at 94 ∘C each for 45 s, 60 ∘C for 45 s and 72 ∘C for 45 s with a final extension at 72 ∘C lasting for 5 min. Following the PCR an aliquot of 20 µL of the PCR bi-PASA product was separated on agarose gel (1.5 %, w : v), which was finally stained with ethidium bromide to permit visualization.
The single-strand conformation polymorphism (SSCP) method was used to identify SNP g.1202T > C by using another primer (SSCP-exon 6); see Table 1. The PCR mixtures contained 0.75 µL of 10 pmol µL−1 forward primer and 0.75 µL of 10 pmol µL−1 reverse primer with the remainder of the mixture as outlined above for the bi-PASA reactions. Aliquots of 2 µL of the PCR products were mixed with 5 µL of denaturing solution containing 98 % (w : v), deionized formamide, 0.01 mM EDTA, 0.25 % (w : v) xylene cyanol, 0.25 % (w : v) bromphenol blue and 2 % (w : v) glycerol; these were heated for 10 min at 98 ∘C and then chilled immediately on ice. Denatured DNA was subjected to electrophoresis on polyacrylamide gel (14 %, w : v) in 1 × TBE buffer at a constant voltage of 200 V overnight at 4 ∘C. Thereafter the gels were stained with 0.36 % (w : v) silver nitrate.
The method described by Pomerantz (1963) was used to measure the activity of tyrosinase in plasma. Briefly, L-dopamine was used as a substrate to measure the activity of the enzyme with a spectrophotometer set at a wavelength of 475 nm. One unit of enzyme was defined as the amount causing the uptake of 1 µL of O2 per minute in a total volume of 3 mL including 1 mg of dopa, 0.1 M sodium phosphate buffer, a pH of 6.8 and a constant temperature of 37 ∘C.
The concentration of plasma phaeomelanin (SpPM) was measured using a spectrophotometric assay (Ozeki et al., 1995, 1996a). An aliquot of 200 µL of each plasma sample was added to 800 µL of phosphate buffer with pH 10.5. The mixture was agitated vigorously for 10 min at room temperature then the mixture was centrifuged at 10 000 rpm for 10 min before 1 mL of chloroform was added to the supernatant to remove fatty impurities. The pale yellow aqueous layer containing phaeomelanin was cleared by centrifugation at 10 000 rpm for 10 min and the absorbance was measured at 400 nm (A400). Plasma concentrations were expressed as the absorbance value A400.
Plasma alkali-soluble melanin (SpASM, comprising phaeomelanin and eumelanin) was measured by the above spectrophotometric analysis, but samples were solubilized using 8M urea–1M sodium hydroxide instead of phosphate buffer. Plasma concentrations were expressed as the value for A400.
Plasma total melanin (SpTM) also was measured by spectrophotometric analysis (Ozeki et al., 1995, 1996a). An aliquot of 100 µL of plasma was mixed with 900 µL of soluene 350 (Packard, Meriden, CT, USA) and then the mixture was heated in a boiling water bath for 45 min. After cooling, the absorbance of the boiled mixture was measured at both 500 nm (A500) and 650 nm (A650) and the SpTM value was taken as the A500 reading. The ratio of the values for A650 : A500 was taken to represent the ratio of eumelanin : total melanin (SpEM : SpTM) in the plasma sample (Ozeki et al., 1996b).
For all plasma samples, duplicate analyses were made.
Functional domain prediction was effected using an online server (http://www.bioinfotllo.org/domac.html). Protein motif prediction was conducted using the PROSITE motif search tool (http://www.cbs.dtu.dk/services/NetNGlyc/). The likelihood of the non-synonymous coding single nucleotide polymorphisms causing a putative functional impact on the protein was predicted using the Protein Variation Effect Analyzer (PROVEAN) software volume 1.1 (http://provean.jcvi.org/seq_submit.php).
Genotypic and allelic frequencies were calculated by direct count. Differences in the distribution of genotypes and alleles between the three groups of sheep and deviation from the Hardy–Weinberg equilibrium were assessed using the χ2 test.
Population genetic diversity parameters, including heterozygosity (He), homozygosity (Ho), effective allele numbers (Ne) and polymorphism content (PIC), were calculated as described by Nei and Li (1979).
The association between genotype and melanin-related traits was analyzed using the general linear model (GLM) procedure (SAS Institute Inc., Cary, NC, USA) according to the following relationship:
where Yij represents the observation of the melanin-related traits, μ is the least squares mean, Pi is the effect of its population (i being either the NPBB, NPN or RM sheep), Gj is the effect of the particular genotype and eij is the random residual.
Values for all traits have been presented as means ± SE (standard errors) of the means.
Statistical significance was determined using the Duncan's new multiple range test and a P value of 0.05 was taken to denote statistical significance.
9.1 Sequence characteristics of TYRP1 and TYRP1 exons
The 4,956bp genomic sequence of the TYRP1 gene of NPBB sheep was assembled and deposited on the GenBank database with accession number EU760771. Eight exons and their partial flanking regions were amplified. Using the GT-AG rule for donor and acceptor sites in the splice sites between exon–intron boundaries (Frandsen et al., 2008) and comparing with sheep TYRP1 gene coding regions (EF102110), the lengths of exons 1–8 for NPBB sheep were respectively 19, 385, 323, 295, 168, 180, 147 and 206 bp. Exon 1 was an un-coding exon so the remaining seven coding exons consisted of an open reading frame comprising 1,614bp (GenBank accession number EU760770) encoding a protein of 537 amino acids with a predicted molecular mass of 60 623 Da (pI 6.48).
Based on the bioinformatics analysis, the signal peptide was identified between amino acids 1 and 23 at the N-terminus. Five N-linked glycosylation sites were retrieved. Sheep TYRP1 has an epidermal growth factor (EGF-like) domain (amino acids 98–1090 and two histidine-rich metal ion-binding sites at amino acids 192–224 and 377–404). A total of 15 cysteine residues were conserved among the tyrosinase gene family (TYRP1, TRYP2 and TYRP; see Fig. 1).
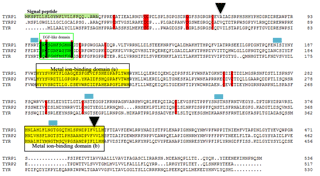
Figure 1Alignment of the sequences for the tyrosinase family of enzymes TYRP1, TYRP2 and TYR. A signal sequence at the N-terminus (residues 1–23) is shaded. The epidermal growth factor (EGF-like) domain and metal ion-binding domains are boxed. The conserved cysteine is marked by a reddish bar. The N-linked glycosylation sites are revealed by the blue boxes. The triangle indicates the non-synonymous mutation. GenBank accession numbers are EU760772, EU760771 and EU760769 for the TYR, TYRP1 and TYRP2, respectively.
9.2 Identification and distribution of TYRP1 gene polymorphisms
The following 12 SNPs were identified by sequencing bi-directionally.
| g.602T > G (intron 1, c.-57T > G) | g.748T > C (exon 2, c.90T > C) |
| g.861C > T (exon 2, c.203C > T) | g.1601T > G (intron 3, c.708 + 15T > G) |
| g.1720A > G (intron 3, c.708 + 134A > G) | g.1742G > A (intron 3, c.708 + 156G > A) |
| g.2066G > T (intron 3, c.709-206G > T) | g.3011T > C (exon 5, c.1038T > C) |
| g.A3449T (intron 5, c.1082-15AA > T) | g.3489T > C (exon 6, c.1107T > C) |
| g.3584T > C (exon 6, c.1202T > C) | g.4760C > A (exon 8, c.1470C > A)* |
| * location corresponded to EU760771 | |
Six of the above sites in genomic location had corresponding locations in
coding regions (EF102110). The sites and their corresponding locations were
as follows.
| g.748T > C | corresponded to c.90T > C |
| g.861C > T | corresponded to c.203C > T |
| g.3011T > C | corresponded to c.1038T > C |
| g.3489T > C | corresponded to c.1107T > C |
| g.3584T > C | corresponded to c.1107T > C |
| g.4760C > A | corresponded to c.1202T > C |
The sites c.90T > C, c.1038T > C, c.1107C > T and c.1407C > A were not changed for encoded amino acids, but the SNPs c.203C > T and c.1202T > C belonged to non-synonymous mutations and led to changes in the amino acids from alanine to valine at position 68 (p.A68V) and valine to alanine at position 401 (p.V401A). Furthermore, the variant p.V401A was clustered into the metal ion-binding domain B. The PROVEAN scores for the mutations p.A68V and p.V401A were 3.234 and −2.624, respectively, raising the possibility that the variation of c.1202T > C (p.A68V) may be deleterious (cutoff = −2.5).
9.3 Genotype and allele frequencies of the TYRP1 gene
The genotypes and allelic frequencies of the two coding SNPs of the TYRP1 gene (c.203C > T and c.1202T > C) in the three groups of sheep are shown in Fig. 2. All groups of sheep exhibited similar genotypes. The χ2 test was used to show that the distributions of genotypes for the TYRP1 gene were in agreement with the Hardy–Weinberg equilibrium in all three groups of sheep. There were marked differences in the distribution of the genotypes between the RM sheep and both groups of sheep from Nanping ( NPBB and NPN sheep). Differences for the NPBB and NPN sheep for the SNPs investigated were not significant.
Values for gene homozygosity (Ho), gene heterozygosity (He), effective allele numbers (Ne) and polymorphism content (PIC) for the above SNPs of the TYRP1 gene are shown in Fig. 3. The values for Ho, He, Ne and PIC for the three groups of sheep were similar. On the basis that a value for PIC between < 0.25 corresponds to a low level of polymorphism and a value between 0.25 and 0.50 corresponds to a medium level of polymorphism (Zhao et al., 2009), the levels of polymorphism for the TYRP1 gene in all three groups of sheep fell in the low to medium range.
The activity of tyrosinase in the plasma of the NPBB sheep was significantly higher (P < 0.01) than in the plasma of the NPN sheep, which had a significantly higher (P < 0.01) activity of plasma tyrosinase than for the RM sheep (Fig. 4a).
Values of SpPM, SpASM and SpTM, as well as the ratio of SpEM : SpTM, were consistently and significantly (P < 0.01) higher for the NPBB than RM sheep. Although the values of SpPM were similar for NPBB and NPN sheep, the concentrations of SpASM, SpTM and the ratio of SpEM : SpTM were significantly lower (P < 0.01) for the NPN than NPBB sheep. Values of SpTM were similar for the NPN and RM sheep but values of SpTM, SpASM and the ratio of SpEM : SpTM were significantly lower (P < 0.01) for the RM than NPN sheep (Fig. 4b).
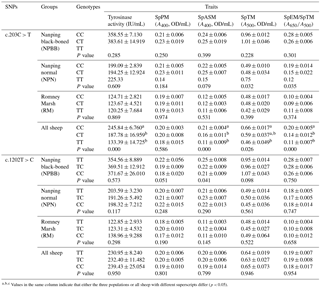
Table 2Association analysis of the TYRP1 gene with black traits (least squares mean value ±1 SE; SE indicates the standard errors of the means) in the three flocks of sheep and all sheep sampled.
Values in the same column indicate that either the three populations or all sheep with different superscripts differ (p < 0.05).
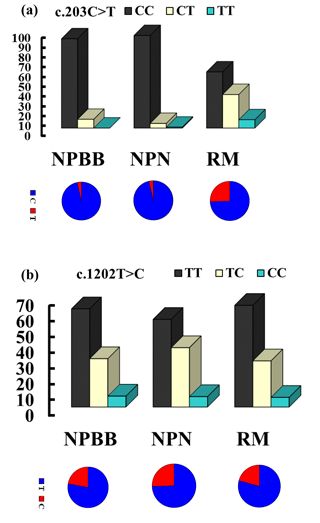
Figure 2The genotype and gene frequencies of the SNPs c.203C > T (a) and c.1202T > C (b) from the TYRP1 gene in Nanping black-boned (NPBB), Nanping normal (NPN) and Romney Marsh (RM) sheep.
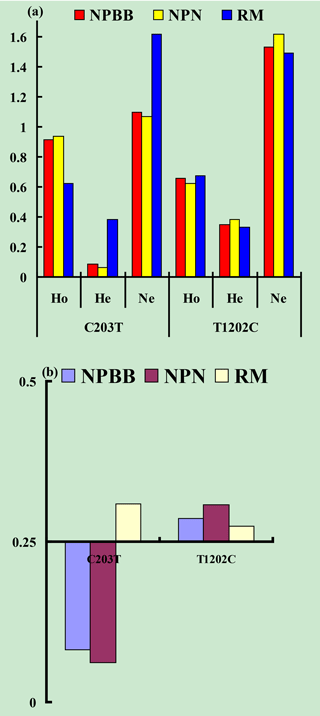
Figure 3Genetic diversity parameters at the SNPs c.203C > T and c.1202T > C from the TYRP1 gene in Nanping black-boned (NPBB), Nanping normal (NPN) and Romney Marsh (RM) sheep. (a) Ho: gene homozygosity; He: gene heterozygosity; Ne: effective allele numbers. (b) PIC: polymorphism information content.
Associations between genotype and melanin-related traits for the SNPs c.203C > T and c.1202T > C are presented in Table 2. In the case of SNP c.203C > T there were no significant (P > 0.05) associations between genotype and melanin-related traits. When the data for all sheep were pooled, in the case of SNP c.203C > T, sheep with the CC genotype showed significantly higher (P < 0.05) values for tyrosinase activity, SpPM, SPTM and the ratio of SpEm : SpTM than sheep with the CT and TT genotypes for which similar values were measured for all traits. In the case of the SNP c.1202T > C, no significant associations were measured for each of the genotypes and melanin-related traits.
In the case of coat color, polymorphism of the site c.203C > T had a significant effect (P < 0.01) on coat color in the NPN group of sheep. No significant effects of polymorphism on coat color was measured for SNP 1202T > C even though the majority of the NPBB sheep of each of the three genotypes had black or brown coats (Table 3).
Tyrosinase is a rate-limiting enzyme for the synthesis of melanin within the enzymatic cascade (Cone et al., 1996; Hall and Orlow, 2005). When the activity of tyrosinase is low, the production of cysteinyl-DOPA occurs without the synthesis of melanin. As the activity of tyrosinase increases cysteinyl-DOPA is oxidized to form phaeomelanin and when the activity of tyrosinase is high the enzymatic cascade tends to result in the production of more eumelanin (Ito et al., 2000). Both TYRP1 and TYRP2 stabilize and improve the activity of tyrosinase.
In the present study, the NPBB sheep had a significantly higher (P < 0.01) activity of tyrosinase in plasma than the NPN and RM sheep. The NPBB sheep also had significantly more (P < 0.01) plasma phaeomelanin, total melanin and alkali-soluble melanin as well as more eumelanin (calculated as SpEM ∕ SpTM × SpTM) than the NPN and RM sheep. The NPBB sheep revealed specific black traits differing from normal sheep and other mammals, confirming the earlier report of a study with fewer NPBB sheep (Deng et al., 2006).
The genomic sequence of 4956 bp, covering eight entire exons and partial flanking regions, of the gene for TYRP1 of the NPBB sheep was retrieved using eight pairs of primers. It was found that there were 12 polymorphic sites including 4 synonymous and 2 non-synonymous mutations (p.A68V and p.V401A) in exonic regions and 6 other sites in intronic regions. In a similar study of pigmentation with Soay sheep, six coding SNPs (including c.90C > T, c.869G > T, c.1107C > T, c.1339A > G, c.1442C > T and c.1470A > C) were identified (16). Three SNPs (viz. c.90C > T, c.1107C > T and c.1479A > C) were shared between the sheep investigated in this study (Nanping indigenous and RM sheep) and the Soay sheep.
Table 3The TYRP1 genotype with coat color distribution in the three groups of sheep.
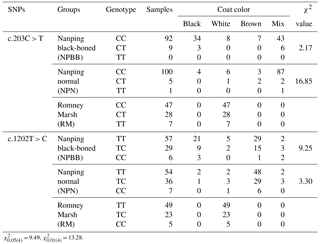
= 9.49, = 13.28.
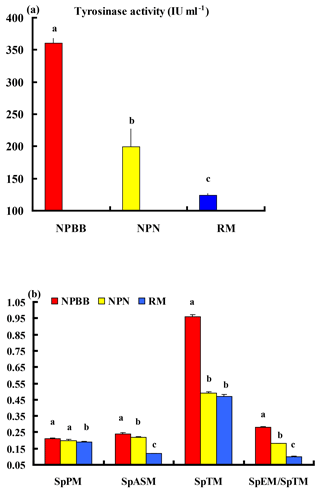
Figure 4Plasma tyrosinase activity (a) and concentrations of phaeomelanin (SpPM), alkali-soluble melanin (SpASM) and total melanin (SpTM) in the plasma as well as the ratio of plasma eumelanin : total melanin (SpEM : SpTM) for the Nanping black-boned, Nanping normal and Romney Marsh (RM) sheep (b). Values presented are means and values shown with different letters (a–c) differ significantly (P < 0.01).
As shown in Fig. 1, the deduced protein TYRP1 was characterized by classical motifs including the signal peptide, an EGF-like domain, conserved cysteine sites, N-linked glycosylation sites and metal ion-binding domains A and B. These motifs are consistent with the sheep family of tyrosinase enzymes having all the important features necessary for folding, maturation, glycosylation and sorting, like active enzymes in the melanosome (Guibert et al., 2004). It is of interest that the variant p.401V > A, which appears to have a deleterious effect on the function of TYRP1, was located at the metal ion-binding domain B.
The results for the bi-PASA analyses for the pooled data for all three groups of sheep in the present study (see Table 2) were interpreted as evidence that for the mutation c.203C > T of the TYRP1 gene, sheep with the CC, compared to those with CT and TT, genotype had significantly higher (P < 0.05) tyrosinase activity in the plasma and plasma contents of alkali-soluble and total melanin as well as ratio of plasma eumelanin : total melanin. However, these differences were not detected within the separate groups of sheep. The great majority of the sheep from the NPBB and NPN groups with CC and CT genotypes displaying the mutation c.203C > T had black, brown or mixed-colored coats. In contrast, within the group of RM sheep all the sheep exhibited white coats. In the cases of sheep displaying the c.1202T > C mutation, there were no significant associations between plasma tyrosinase and melanin for the three genotypes (Table 2). In these groups of sheep the majority of NPBB and NPN sheep with the TT and TC genotypes had black, brown or mixed-colored coats, whereas all the RM sheep had white coats (Table 3). Overall, these observations are considered to be consistent with the previous report that plasma tyrosinase activity is higher in sheep with black skin than in those with white skin (Pomerantz and Ances, 1975; Iwata et al., 1990). In Soay sheep the coat is either dark brown or a light tawny color. A non-synonymous mutation (c.869G > T; p.290C > F) of the TYRP1 gene has been reported to be responsible for the light phenotype (Gratten et al., 2007). This site mutation has also been reported to have an association with partial dominance for body size in Soay sheep (Gratten et al., 2008).
Recently the gene expression profiles in sheep with either white or black coat colors were compared using a transcriptome sequencing technique. A total of 2235 known genes and 845 novel genes were found to be expressed in sheep with black compared to white skin. The TYRP1 gene was shown to have the highest level of differential expression. There were 13 coat color genes, including the tyrosinase gene family, expressed higher in sheep with black compared to white skin (Fan et al., 2013). Recently in a study with NPBB and NPN sheep, no differences for the mRNA expression levels were found for the TYRP1 gene in different organs and tissues (Deng WD, unpublished data). It appears that coat color in the sheep is influenced by several genes, pathways or networks (Bellei et al., 2011). Indeed, it is known that more than 150 genes and 300 genetic loci (Norris and Whan, 2008; Hofreiter and Schöneberg, 2010; Cieslak et al., 2011; Aydin et al., 2012; Li et al., 2014) and environmental factors affect the pigmentation of the integument, including coat color, in animals. Given the latter, coat color in NPBB is likely to be affected by more than the variants of the TYRP1 gene.
It is concluded that while it is clear that NPBB sheep have higher contents of melanin and related pigments in tissues and organs than NPN and RM sheep, these traits were not solely due to variations in the coding SNPs of the TYRP1 gene c.203C > T and c.1202T > C. Further studies using whole genome re-sequencing technology should be undertaken to determine the genes responsible for the black traits in the novel and economically important NPBB sheep.
Data are available upon reasonable request from the corresponding author.
The authors declare that they have no conflict of interest.
This work was supported by grants from the National Nature Science
Foundation of China (31660634) and Yunnan Province (2016FA007).
The authors appreciate the help of the staff from the Animal Science and
Veterinary Bureau in Nanping and Jianchuan counties and local farmers
Fuhai Yang and Sinanduojie during sample collection.
Edited by: Steffen Maak
Reviewed by: two anonymous referees
Aliev, G. A., Rachkovskii, M. L., and Krymova, M. A.: Morpho-physiological study of the inhibitory effect of modifier genes on melanogenesis in the ontogenesis of sheep of the Tadzhik breed, Genetika, 23, 1645–1657, 1987.
April, C. S., Jackson, I. J., and Kidson, S. H.: The cloning and sequencing of a cDNA coding for chick tyrosine-related protein-1, Biochim. Biophys. Acta, 1395, 7–12, 1998.
Aydin, I. T., Hummler, E., Smit, N. P., and Beermann, F.: Coat color dilution in mice because of inactivation of the melanoma antigen MART-1, Pigment Cell Melanoma Res., 25, 37–46, https://doi.org/10.1186/1471-2164-14-389, 2012.
Becker, D., Otto, M., Ammann, P., Keller, I., Drögemüller, C., and Leeb, T.: The brown coat colour of Coppernecked goats is associated with a non-synonymous variant at the TYRP1 locus on chromosome 8, Anim. Genet., 46, 50–54, https://doi.org/10.1111/age.12240, 2015.
Bellei, B., Pitisci, A., Catricalà, C., Larue, L., and Picardo, M.: Wnt/β-catenin signaling is stimulated by α-melanocyte-stimulating hormone in melanoma and melanocyte cells: implication in cell differentiation, Pigment Cell Melanoma Res., 24, 309–325, https://doi.org/10.1111/j.1755-148X.2010.00800.x, 2011.
Berryere, T. G., Schmutz, S. M., Schimpf, R. J., Cowan, C. M., and Potter, J.: TYRP1 is associated with dun coat colour in Dexter cattle or how now brown cow, Anim. Genet., 34, 169–175, https://doi.org/10.1046/j.1365-2052.2003.00985.x, 2003.
Cieslak, M., Reissmann, M., Hofreiter, M., and Ludwig, A.: Colours of domestication, Biol. Rev. Camb. Philos. Soc., 86, 885–899, https://doi.org/10.1111/j.1469-185X.2011.00177.x, 2011.
Cone, R. D., Lu, D., Koppula, S., Vage, D. I., Klungland, H., Boston, B., Chen, W., Orth, D. N., Pouton, C., and Kesterson, R. A.: The melanocortin receptors: agonist, antagonists, and the hormonal control of pigmentation, Recent Prog. Horm. Res., 51, 287–318, 1996.
de Araújo Lima, F., de Toledo Gonçalves, F., and Fridman, C.: SLC24A5 and ASIP as phenotypic predictors in Brazilian population for forensic purposes, Leg. Med. (Tokyo), 17, 261–266, https://doi.org/10.1016/j.legalmed.2015.03.001, 2015.
Deng, W. D., Yang, S. L., Huo, Y. Q., Gou, X., Shi, X. W., and Mao, H. M.: Physiological and genetic characteristics of black-boned sheep (Ovis aries), Anim. Genet., 37, 586–588, https://doi.org/10.1111/j.1365-2052.2006.01530.x, 2006.
Deng, W. D., Xi, D. M., Gou, X., Yang, S. L., Shi, X. W., and Mao, H. M.: Pigmentation in Black-boned sheep (Ovisaries): association with polymorphism of the Tryosinase gene, Mol. Biol. Rep., 35, 379–385, https://doi.org/10.1007/s11033-007-9097-z, 2008.
Dubey, S. and Roulin, A.: Evolutionary and biomedical consequences of internal melanins, Pigment Cell Melanoma Res., 27, 327–338, https://doi.org/10.1111/pcmr.12231, 2014.
Fan, R., Xie, J., Bai, J., Wang, H., Tian, X., Bai, R., Jia, X., Yang, L., Song, Y., Herrid, M., Gao, W., He, X., Yao, J., Smith, G. W., and Dong, C.: Skin transcriptome profiles associated with coat color in sheep, BMC Genomics., 14, 389, https://doi.org/10.1186/1471-2164-14-389, 2013.
Frandsen, P. M., Madsen, L. B., Bendixen, C., and Larsen, K.: Porcine gamma-synuclein: molecular cloning, expression analysis, chromosomal localization and functional expression, Mol. Biol. Rep., 36, 971–979, https://doi.org/10.1007/s11033-008-9270-z, 2008.
Gratten, J., Beraldi, D., Lowder, B. V., McRae, A. F., Visscher, P. M., Pemberton, J. M., and Slate, J.: Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep, Proc. Biol. Sci., 274, 619–626, https://doi.org/10.1098/rspb.2006.3762, 2007.
Gratten, J., Wilson, A. J., McRae, A. F., Beraldi, D., Visscher, P. M., Pemberton, J. M., and Slate, J.: A localized negative genetic correlation constrains microevolution of coat color in wild sheep, Science, 319, 318–320, 2008.
Guibert, S., Girardot, M., Leveziel, H., Julien, R., and Oulmouden, A.: Pheomelanin coat colour dilution in French cattle breeds is not correlated with the TYR, TYRP1 and DCT transcription levels, Pigment Cell Res., 17, 337–345, https://doi.org/10.1111/j.1600-0749.2004.00152.x, 2004.
Hall, A. M. and Orlow, S. J.: Degradation of tyrosinase induced by phenylthiourea occurs following Golgi maturation, Pigment Cell Res., 18, 122–129, https://doi.org/10.1111/j.1600-0749.2005.00213.x, 2005.
Hofreiter, M. and Schöneberg, T.: The genetic and evolutionary basis of colour variation in vertebrates, Cell Mol. Life Sci., 67, 2591–2603, https://doi.org/10.1007/s00018-010-0333-7, 2010.
Ito, S. and Wakamatsu, K.: Chemistry of mixed melanogenesis–pivotal roles of dopaquinone, Photochem. Photobiol., 84, 582–592, https://doi.org/10.1111/j.1751-1097.2007.00238.x, 2008.
Ito, S., Wakamatsu, K., and Ozeki, H.: Chemical analysis of melanins and its application to the study of the regulation of melanogenesis, Pigment Cell Res., 13, 103–109, https://doi.org/10.1034/j.1600-0749.13.s8.19.x, 2000.
Iwata, M., Corn, T., Iwata, S., Everett, M. A., and Fuller, B. B.: The relationship between tyrosinase activity and skin color in human foreskins, J. Invest. Dermatol., 95, 9–15, https://doi.org/10.1111/1523-1747.ep1287267, 1990.
Li, M. H., Tiirikka, T., and Kantanen, J.: A genome-wide scan study identifies a single nucleotide substitution in ASIP associated with white verus non-white coat-colour variation in sheep (Ovis aries), Heredity (Edinb), 112, 122–131, 2014.
Liu, Q., Thorland, E. C., Heit, J. A., and Sommer, S. S.: Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes, Genome Res., 7, 389–398, https://doi.org/10.1101/gr.7.4.389, 1997.
Lyons, L. A., Foe, I. T., Rah, H. C., and Grahn, R. A.: Chocolate coated cats: TYRP1 mutations for brown color in domestic cats, Mamm. Genome., 16, 356–366, https://doi.org/10.1007/s00335-004-2455-4, 2005.
Manga, P., Sato, K., Ye, L., Beermann, F., Lamoreux, M. L., and Orlow, S. J.: Mutational analysis of the modulation of tyrosinase by tyrosinase-related proteins 1 and 2 in vitro, Pigment Cell Res., 13, 364–374, https://doi.org/10.1034/j.1600-0749.2000.130510.x, 2000.
Nadeau, N. J., Mundy, N. I., Gourichon, D., and Minvielle, F.: Association of a single-nucleotide substitution in TYRP1 with roux in Japanese quail (Coturnix japonica), Anim. Genet., 38, 609–613, https://doi.org/10.1111/j.1365-2052.2007.01667.x, 2007.
Nei, M. and Li, W. H.: Mathematical model for studying genetic variation in terms of restriction endonucleaes, P. Natl. Acad. Sci. USA., 76, 5269–5273, https://doi.org/10.1073/pnas.76.10.5269, 1979.
Norris, B. J. and Whan, V. A.: A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep, Genome Res., 18, 1282–1293, https://doi.org/10.1101/gr.0090.107, 2008.
Okumura, N., Hayashi, T., Sekikawa, H., Matsumoto, T., Mikawa, A., Hamasima, N., and Awata, T.: Sequences and mapping of genes encoding porcine tyrosinase (TYR) and tyrosinase-related proteins (TYRP1 and TYRP2), Anim. Genet., 36, 513–516, https://doi.org/10.1111/j.1365-2052.2005.01353.x, 2005.
Okumura, N., Hamasima, N., Suzuki, K., and Awata, T.: Genomic organization of the porcine dopachrome tautomerase (DCT) gene, Anim. Sci. J., 79, 182–186, https://doi.org/10.1111/j.1740-0929.2008.00515.x, 2008.
Ozeki, H., Ito, S., Wakamatus, K., and Hirobe, T.: Chemical characterization of hair melanins in various coat-color mutants of mice, J. Invest. Dermatol., 105, 361–366, https://doi.org/10.1111/1523-1747.ep12320792, 1995.
Ozeki, H., Ito, S., and Wakamatus, K.: Chemical characterization of melanins in sheep wool and human hair, Pigment Cell Res., 9, 51–57, https://doi.org/10.1111/j.1600-0749.1996.tb00088.x, 1996a.
Ozeki, H., Ito, S., Wakamatus, K., and Thody, A. J.: Spectrophotometric characterization of eumelanin and pheomelanin in hair, Pigment Cell Res., 9, 265–270, https://doi.org/10.1111/j.1600-0749.1996.tb00116.x, 1996b.
Pomerantz, S. H.: Separation, purification, and properties of two tyrosinases from hamster melanoma, J. Biol. Chem., 238, 2351–2357, 1963.
Pomerantz, S. H. and Ances, I. G.: Tyrosinase activity in human skin. Influence of race and age in newborns, J. Clin. Invest., 55, 1127–1131, https://doi.org/10.1172/JCI108014, 1975.
Rieder, S., Taourit, S., Mariat, D., Langlois, B., and Guérin, G.: Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equuscaballus), Mamm. Genome., 12, 450–455, https://doi.org/10.1007/s003350020017, 2001.
Rooryck, C., Roudaut, C., Robine, E., Musebeck, J., and Arveiler, B.: Oculocutaneous albinism with TYRP1 gene mutations in a Caucasian patient, Pigment Cell Res., 19, 239–242, https://doi.org/10.1111/j.1600-0749.2006.00298.x, 2006.
Schmidt-Kuntzel, A., Eizirik, E., O'Brien, S. J., and Menotti-Raymond, M.: Tyrosinase and tyrosinase related protein I alleles specify domestic cat coat color phenotypes of the albino and brown loci, J. Hered., 96, 289–301, https://doi.org/10.1093/jhered/esi066, 2005.
Schmutz, S. M., Berryere, T. G., and Goldfinch, A. D.: TYRP1 and MC1R genotypes and their effects on coat color in dogs, Mamm. Genome., 13, 380–387, https://doi.org/10.1007/s00335-001-2147-2, 2002.
Slominski, A., Tobin, D. J., Shibahara, S., and Wortsman, J.: Melanin pigmentation in mammalian skin and its hormonal regulation, Physiol. Rev., 84, 1155–1228, https://doi.org/10.1152/physrev.0004 4.2003, 2004.
Sturm, R. A., O'Sullivan, B. J., Box, N. F., Smith, A. G., Smit, S. E., Puttick, E. R., Parsons, P. G., and Dunn, I. S.: Chromosomal structure of the human TYRP1 and TYRP2 loci and comparison of the tyrosinase-related protein gene family, Genomics, 29, 24–34, https://doi.org/10.1006/geno.1995.1211, 1995.
Toyoda, R., Kasai, A., Sato, S., Wada, S., Saiga, H., Ikeo, K., Gojobori, T., Numakunai, T., and Yamamoto, H.: Pigment cell lineage-specific expression activity of the ascidian tyrosinase-related gene, Gene, 332, 61–69, https://doi.org/10.1016/j.gene.2004.01.030, 2004.
Utzeri, V. J., Ribani, A., and Fontanesi, L.: A premature stop codon in the TYRP1 gene is associated with brown coat colour in the European rabbit (Oryctolaguscuniculus), Anim. Genet., 45, 600–603, https://doi.org/10.1111/age.12171, 2014.
Vachtenheim, J. and Borovanský, J.: “Transcription physiology” of pigment formation in melanocytes: central role of MITF, Exp. Dermatol., 19, 617–627, https://doi.org/10.1111/j.1600-0625.2009.01053.x, 2010.
Xu, Y., Zhang, X. H., and Pang, Y. Z.: Association of tyrosinase (TYR) and tyrosinase-related protein 1 (TYRP1) with melanic plumage color in Korean quails (Coturnixcoturnix), Asian-Australas, J. Anim. Sci., 26, 1518–1522, https://doi.org/10.5713/ajas.2013.13162, 2013.
Zdarsky, E., Favor, J., and Jackson, I. J.: The molecular basis of brown, an old mouse mutation, and of an induced revertant to wild type, Genetics, 126, 443–449, 1990.
Zhao, M., Chen, H., Wang, X., Yu, H., Wang, M., Wang, J., Lan, X. Y., Zhang, C. F., Zhang, L. Z., Guo, Y. K., Zhang, B., and Hu, S. R.: aPCR-SSCP and DNA sequencing detecting two silent SNPs at KAP8.1 gene in the cashmere goat, Mol. Biol. Rep., 36, 1387–1391, https://doi.org/10.1007/s11033-008-9325-1, 2009.
- Abstract
- Introduction
- Materials and methods
- Sheep and collection of blood
- Isolation of exons and identification of DNA polymorphisms
- Genotyping
- Measurement of melanin-related traits
- Bioinformatics analyses
- Genetic diversity and statistical analyses
- Results
- Association analysis of the TRYP1 genotype with melanin-related traits and coat color
- Discussion
- Data availability
- Competing interests
- Acknowledgements
- References
- Abstract
- Introduction
- Materials and methods
- Sheep and collection of blood
- Isolation of exons and identification of DNA polymorphisms
- Genotyping
- Measurement of melanin-related traits
- Bioinformatics analyses
- Genetic diversity and statistical analyses
- Results
- Association analysis of the TRYP1 genotype with melanin-related traits and coat color
- Discussion
- Data availability
- Competing interests
- Acknowledgements
- References