the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The association of RNF34 and RNF128 with carcass and meat quality traits of Chinese Simmental-cross steers
Jun-Zheng Zhang
Azher Nawaz
Si-Han Wang
Quan Tian
Chun-Yin Geng
Ying Hai-Jin
Shuang Ji
In Chinese Simmental-cross steers, carcass and meat quality traits are investigated by identifying SNPs (single-nucleotide polymorphisms) in the RNF34 and RNF128 genes. Statistical analysis showed that for 3′ UTR-588 G>A, a SNP of RNF34 was significantly associated with kidney weight, testis weight, and tare weight (P<0.05), and for I1-2380C>T, a SNP of RNF128 was significantly related to forepaw weight, dressed weight, carcass brisket depth, carcass depth, and hind leg length (P<0.05). This study highlights the importance of polymorphism, suggesting that RNF34 and RNF128 polymorphisms could be important genetic factors that affect carcass and meat quality in beef cattle. Therefore, in beef cattle production and breeding, these SNPs might be valuable marker in future marker-assisted selection programs to determine meat quality traits. However, validation in a larger sample size of the Simmental-cross steers is necessary to verify these findings across a broader population.
Background: This research aims to explore the potential influence of really interesting new gene (RING) finger protein (RNF34) 3′ UTR-588G>A and RING finger protein (RNF128) I1-2380C>T polymorphisms associated with carcass and meat quality traits in Chinese Simmental-cross steers. Methods: Restriction enzyme digestion and sequencing is employed to detect genotypes of RNF34 3′ UTR-588 G>A and RNF128 I1-2380C>T in Chinese Simmental-cross steers. Then, the association of novel single-nucleotide polymorphisms in the 3′ UTR region of RNF34 and the intron regions of the RNF128 gene is analyzed to determine the meat quality traits of the animals. Results: This study revealed a significant association between RNF34 3′ UTR-588 G>A and kidney weight, testis weight, and tare weight (P<0.05). Furthermore, the RNF128 I1-2380C>T variant exhibited a significant link to multiple carcass measurements, indicating its potential association with forepaw weight, dressed weight, carcass depth, hind leg length, and carcass brisket depth (P<0.05). Conclusion: This study highlights the importance of genetic factors that link the variations in RNF34 and RNF128 and their influence on carcass and meat quality in beef cattle. Thus, these variants might be novel valuable markers for meat quality traits in future marker-assisted selection programs in beef cattle breeding and production.
- Article
(1037 KB) - Full-text XML
- BibTeX
- EndNote
With the advancements in molecular biology techniques, the identification of single-nucleotide polymorphisms (SNPs) has been widely used to study the effects of genetic mutations on animal performance, which could be used for a molecular marker-assisted approach to breeding and production (Reshma and Das, 2021; Aly and Sabri, 2015; Das et al., 2021; Deng et al., 2023; Ye et al., 2022; Nawaz et al., 2024). RING (really interesting new gene) finger proteins, RNF34 and RNF128 play an important role in many biological processes, such as protein degradation; protein–protein interaction; and cellular pathways such as signal transduction, cell cycle regulation, DNA repair, inflammatory response, and apoptosis (Cai et al., 2022b). Several papers showed that E3 ubiquitin-protein ligases are master regulators of adaptive thermogenesis and energy metabolism in brown fat cells. The RNF34 is a bona fide E3 ubiquitin-protein ligase for the peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) and negatively regulates brown fat cell metabolism (Sheng et al., 2023).
RNF34 binds to the PGC-1α at its C-terminal region and targets it for degradation independently of the previously identified N-terminal phosphor-degron motif (Liu et al., 2023; Wei et al., 2012; Liu et al., 2008b). Deactivating RNF34 within brown fat cells disrupts its interaction with PGC-1α protein levels and subsequently upregulates UCP1 (uncoupling protein 1), ultimately resulting in increased oxygen consumption (Qi and Ding, 2012; Wang and Deng, 2011b). Nevertheless, the contrary effects are observed in brown fat cells with ectopic expression of wild-type RNF34 instead of its ligase activity-defective mutant (Heissmeyer and Rao, 2004). Interestingly, β3-adrenergic receptor signaling and cold exposure, conditions that trigger expression of PGC-1α expression, suppress RNF34 expression in brown fat cells. This reveals a dynamic interplay between E3 ligase for PGC-1α, RNF34, and thermogenesis, suggesting its physiological importance in heat production regulation (Janowski et al., 2012; Schartner et al., 2009b).
The RNF128 gene participates in cellular processes such as cell cycle regulation, DNA repair, inflammatory response, and apoptosis (Schartner et al., 2009b; Bai et al., 2020). The activation of the T cell is tightly regulated to avoid autoimmunity. The GRAIL protein, associated with energy metabolism in T cells, is encoded by RNF128 and an E3 ubiquitin-protein ligase associated with T cell tolerance. Intriguingly, degradation and ubiquitination of CD40L by RNF128 are one cause of T cell incompetence (Nurieva et al., 2010; Anandasabapathy et al., 2003). In the latest research, it has been found that the expressions of RNF128 and RNF34 displayed a substantial difference between younger and adult cattle (Liu et al., 2008b).
The selection of the RNF34 and RNF128 genes in this study was based on their functionality regarding metabolic pathways, with broader effects that have potentially indirect implications for carcass and meat quality traits in cattle. Although RNF34 is principally associated with proteolytic degradation via its function as an E3 ubiquitin-protein ligase and RNF128 appears to be a regulator of some immune response T cell activation genes, these quantitative trait loci (QTLs) may also influence metabolism for energy production under heat or oxidative stress conditions that affect muscle (energy content) and fat deposition in livestock (Sheng et al., 2023; Cai et al., 2022a).
Moreover, genomic studies in other species and whole-genome research often imply that genes involved in the ubiquitination pathway and inflammation response signaling pathways may have an indirect effect on growth-related meat quality (Deng et al., 2024; Leal-Gutiérrez et al., 2018). Evidence in support of this hypothesis is the identification through QTL mapping and GWAS of regions that overlap or reside near RNF34 and RNF128 with associations to economically relevant traits, including those in cattle and other livestock species. Although its direct functions do not suggest an apparent association, these reports together give us the reason to explore RNF34 and/or RNF128 as candidate genes for carcass meat quality traits. It seems that while there is substantial research related to QTL mapping, GWAS, and candidate genes for economically relevant traits, specific evidence for RNF34 and RNF128 in relation to carcass meat quality traits in livestock has not yet been reported in the available literature.
To further substantiate the relationship of RNF34 and RNF128 with beef quality traits, we scrutinize the appropriate QTL (quantitative trait locus) and GWAS (genome-wide association study) literature. QTL mapping in cattle has been used to identify various chromosomal regions correlated with carcass weight, fat deposition, and muscle development. Some of these regions overlap with the chromosomal positions of RNF34 and RNF128, which found that these genes are located in regions associated with carcass and meat quality traits. The fact that RNF34 and RNF128 genes are located near these regions indicates that these genes may also affect meat quality, thereby appearing in the list of candidate genes for marker-assisted selection in beef cattle breeding. Additionally, comparative studies in other species, such as pigs and chickens, have shown that genes involved in the ubiquitin-proteasome pathway, such as RNF34, are associated with traits such as muscle fiber development and fat deposition (Antonaros et al., 2019a; Sanglard et al., 2023a). These findings suggest that RNF34 and RNF128 could play a similar role in cattle, potentially influencing meat quality through their effects on cellular metabolism and tissue differentiation. This study aims to generate potential evidence to incorporate marker-assisted selection into breeding strategies such as crossbreeding and pure breeding and the vital effort to preserve important genetic diversity.
Therefore, RNF34 and RNF128 may be candidate genes influencing meat quality and carcass of beef cattle (Afonso et al., 2020). At present, only a limited information is available on the genetic polymorphism of RNF34 and RNF128 genes in cattle, and the effect of the genetic variants of these genes remains indecisive. The main objective of the study is to examine the SNPs in these two genes and finding their association with carcass and meat quality traits in Chinese Simmental-cross steers. SNPs in these genes were determined by restriction endonuclease analysis (REA) and polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) as these are cost-effective and simple techniques which recognize the specific DNA sequence by chosen restriction enzymes that cut the DNA at specific restriction sites to genotype the known polymorphisms (Hashim and Al-Shuhaib, 2019; Antonaros et al., 2019b; Sanglard et al., 2023b).
2.1 Experimental materials
In this study, a total of 255 Chinese Simmental-cross steers that are 28 months of age were selected from the Baolongshan cattle farm in Inner Mongolia. The selection process involved choosing 21 bulls and 2000 female cows from the offspring of Simmental cattle. However, instead of a purely random selection, the animals were chosen based on a controlled breeding program designed to maintain genetic diversity while ensuring a representative sample of the population. The breeding program involved crosses between purebred Simmental sires and dams of known pedigree, which allowed us to account for the genetic relationships among the animals. This approach minimized potential confounding effects in the genetic association analysis by reducing the impact of population structure and relatedness.
Blood samples (10 mL) from the jugular vein were collected in acid citrate dextrose (ACD), as anticoagulant. Natural samples after the treatment were kept frozen at −80 °C (Tian et al., 2013; Xiao et al., 2016). In total, 1 mL out of all of the blood was taken for DNA extraction under the manufacturer's protocol using a DNA extraction kit (Tiangen, Beijing, China).
2.2 Experimental methods
2.2.1 Slaughter process
The slaughtering was performed according to established standard protocols in order to safeguard the welfare of each individual and ensure a certain level of quality within data collection. To minimize stress that could as well affect meat quality traits, all the 255 Chinese Simmental-cross steers were transported under controlled conditions to the slaughter plant. Animals were allowed 24 h rest with water but no feed (to reduce gut content) on arrival at the facility. Each cow's head was secured within the dizzying box to prevent movement. A pneumatic dizzying device was aimed precisely at the intersection of the cow's two horns and the diagonal of both eyes. The device was quickly activated to render the cow unconscious. The slaughtering process started with a stunning that was done by a captive bolt pistol for fast and humane euthanasia. The animals were then exsanguinated by severing the major blood vessels in the neck, ensuring a complete and rapid bleed-out, which is critical for both animal welfare and meat quality. After exsanguination, the carcasses were hung on a rail system and moved through the slaughter line.
The carcasses were then skinned, eviscerated, and split into halves. During evisceration, the visceral organs were carefully removed and weighed as part of the trait measurements. The carcasses were washed and immediately moved into a refrigerated room set to between 0 and 4° C, where they were kept for 24 h before further processing and trait measurement. This chilling step is essential to reduce the risk of microbial contamination and to stabilize the carcass for accurate measurement of meat quality traits.
2.2.2 Trait measurements
In this study, a total of 42 traits were measured, as shown in Table 1. Before trait measurements, all the carcasses were preserved in refrigerated rooms at 0 to 4 °C for 24 h. Trait were measured on the basis of cutting standards (GB/T17238-1998) for chilled and fresh beef of China (China Standard Publishing House) (Tian et al., 2013). The final weight of the body, living quality index of beef (QIB), and rib eye area (by ultrasound) were noted before slaughter. All visceral indicators were weighed after slaughter, including the spleen, large intestine, small intestine, heart, liver, kidney, and fat belly (Guo et al., 2016; Tian et al., 2013). Other carcass properties were also recorded, including the carcass weight, slaughter rate, bone weight, head weight, tare weight, hind leg circumference, fat color score, hind leg width, and carcass brisket depth. The described measurements were determined strictly according to established measurement standards (Li et al., 2017).
Table 1The number of records and mean and standard errors for traits included in the association analyses.
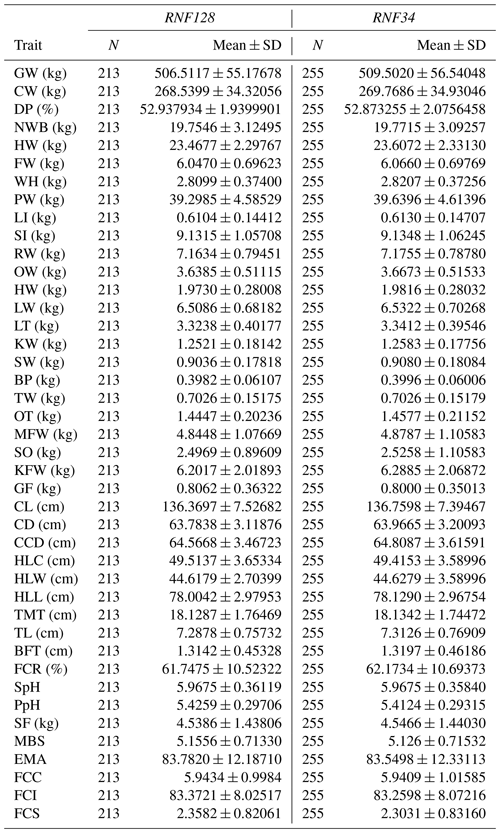
GW: gross weight; CW: carcass weight; DP: dressing percentage; NWB: net weight of bone; HW: head weight; FW: forepaw weight; WH: Weight heels; LI: large intestine; SI: small intestine; RW: rumen weight; OW: omasum weight; HW: heart weight; LW: liver weight; LT: lung, trachea; KW: kidney weight; SW: spleen weight; BP: bull penis; TW: testes weight; OT: ox tail; MFW: mesenteric fat weight; SO: stomach oil; KFW: kidney fat weight; GF: genital fat; CL: carcass length; CD: carcass depth; CCD: carcass chest depth; HLC: hind leg circumference; HLW: hind leg width; HLL: hind leg length; TMT: thigh meat thickness; TL: thickness of loin; BFT: back fat thickness; FCR: fat coverage rate of carcass; SpH: slaughter pH; SF: shearing force; MBS: marbling score; EMA: eye muscle area; SE: standard error of mean.
2.2.3 Ultrasound device and procedure
An Aloka SSD-500V ultrasound device with a 3.5 MHz linear array transducer was used to assess the rib eye area in live beef cattle. The transducer was inserted between the 12th and 13th rib sites, and repeated measurements were carried out to ensure accuracy. The images were analyzed using BIA Pro software, and the rib eye area was measured by detecting muscle boundaries without fat or bone structures.
2.2.4 Color score assessment
The color of fat and meat was scored by adhibition equipment Olympic Minolta CR-400 chromameter. A white calibration plate (Minolta CR-A43) was used to calibrate the device prior to each session for reliable measurements (Konica Minolta Sensing, Inc., 1998). The color scores were obtained by taking readings from three different locations on the longissimus dorsi muscle for meat color and subcutaneous fat for fat color. The device provides L* (lightness), a* (redness), and b* (yellowness) values, which are then converted into a standardized color score using the CIELAB color space method.
2.2.5 Primers designing and PCR amplification
Primer 5 software was used to design primers targeting bovine RNF34 (NC_037344.1) and RNF128 (NC_037357.1) sequences, mentioned in Table 2, while genotyping primer pairs were synthesized by Sangon Biotech (Shanghai, China). The gene-specific primer pairs are RNF34 forward: 5′-CGGGCTGTTTCCCAGGTTCT-3′, RNF34 reverse: 5′-CCCAATGATGTTGAAACGCAGA-3′, RNF128 forward: 5′-GAGCAAACAGAGGCTTACACAAC-3′, and RNF128 reverse: 5′-TCAGTCTTACCTCTTTGCCACTAG-3′.
A reaction mixture of total 20 µL volume was prepared for a polymerase chain reaction: bovine genomic DNA (100 ng), each primer (10 pmol L−1), deoxynucleotide triphosphates (dNTP) (200 µmol L−1 of each), MgCl2 (1.5 mmol L−1), 2.5 µL buffer (10× concentrate: 200 mmol L−1 Tris-HCl, pH 8.4, 500 mmol L−1 KCl), and 1.0 unit of Taq DNA polymerase. The following cycling conditions were given for PCR amplification (Xiao et al., 2016): 95 °C for 5 min (initial denaturation) followed by 30 cycles of 95 °C for 30 s (denaturation), 60 °C for 30 s (annealing), and 72 °C for 40 s (extension), with a final extension step at 72 °C for 10 min, as shown in Fig. 1.
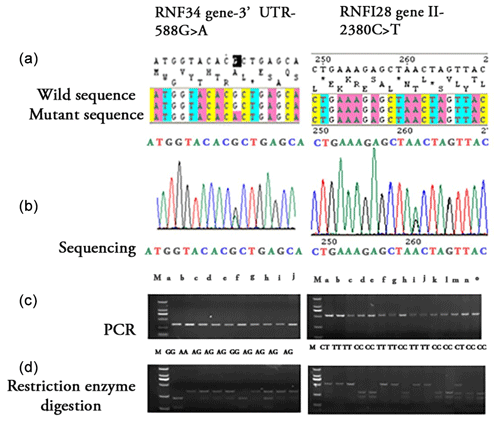
Figure 1Identification of bovine RNF34 gene 3′ UTR-588G>A and RNF128 gene I1-2380C>T by sequencing and restriction enzyme digestion analysis of PCR products. (a) Wild-type and mutant sequences of the RNF34 3′ UTR-588G>A and RNF128 I1-2380C>T genes. The sequences shown highlight the nucleotide changes from G to A (RNF34) and C to T (RNF128). (b) Sequencing chromatograms showing the SNP locations for RNF34 3′UTR-588G>A and RNF128 I1-2380C>T. The chromatograms illustrate the nucleotide variations at positions 588 and 2380, respectively. (c) PCR amplification results displaying the genotypes for the RNF34 and RNF128 SNPs. Lanes are labeled according to the genotype of the individual animals, with M indicating the DNA marker. (d) Restriction enzyme digestion analysis of the PCR products for RNF34 and RNF128. The digested products are shown for each genotype, demonstrating the presence or absence of the restriction site corresponding to the SNP.
2.2.6 Genomic DNA extraction
Genomic DNA was extracted from the collected blood samples using a Tiangen DNA extraction kit (Tiangen, Beijing, China) according to the manufacturer's protocol. Briefly, 1 mL of whole blood was mixed with a lysis buffer and proteinase K, which was followed by incubation at 56 °C until complete lysis. The samples were then subjected to ethanol precipitation, and the DNA was purified using a silica-based spin column. Concentration and quality of the DNA were determined using a NanoDrop (Thermo Fisher Scientific, USA) spectrophotometer and agarose gel electrophoresis. The DNA extracts were then kept frozen at −20 °C until further use.
2.2.7 Development of SNP library
A SNP library was created for the analysis of polymorphisms that exist within two putative target genes, RNF34 and RNF128. The target regions, the 3′ UTR of RNF34 and intron 1 of RNF128 were amplified with specific primers designed on reference sequences from NCBI. All sequences were confirmed by Sanger sequencing of purified PCR products using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The sequencing data were analyzed using the BioEdit software to search for single-nucleotide polymorphisms within the target regions. The identified SNP were subjected for association analysis with carcass and meat quality traits.
2.2.8 Selection of gene regions
This study focuses on the 3′ UTR of RNF34 and intron 1 of RNF128 due to their potential to harbor SNPs that affect gene expression (Liu et al., 2008a; Wang and Deng, 2011a). The 3′ UTR regulates mRNA stability and protein levels, directly influencing traits like carcass and meat quality (Schartner et al., 2009a; Petroski and Deshaies, 2005a). Focusing on these regions allowed for a more efficient and cost-effective investigation of genetic variations linked to these traits.
2.2.9 SNP detection and genotyping
Two polymorphisms in RNF34 and RNF128 genes were identified through the sequencing and confirmed through REA (restriction endonuclease analysis) using enzymes as restriction endonuclease (Bpu1102 and Spe I, New England Biolabs, MA, USA). Bpu1102 was used to distinguish the genotypes of the PCR products of the RNF34 gene while Spe I used for RNF128 gene. Restriction digestion of the amplicons of the SNPs of two genes was conducted at 37 °C for 6 h – 18 µL, final volumes with PCR product (8 µL), enzyme buffer (1.8 µL), and restriction enzyme (0.4 µL). The products of PCR digestion of both genes were resolved and visualized on 3 % agarose gels to distinguish the bands representing three different genotypes (Tian et al., 2013; Xiao et al., 2016; Li et al., 2017).
2.2.10 Statistical analysis
Allelic and genotypic frequencies for polymorphism of each gene were determined based on its information content to understand the diversity in the studied population. Genotype frequencies were calculated and analyzed by significance testing. The genotypic effects of the RNF34 gene and RNF128 gene were determined using the general linear model (GLM) and one-way ANOVA using SPSS (version 13.0) (Korkmaz-Ağaoğlu et al., 2019).
3.1 PCR amplification
In total, genomic DNA from 255 Simmental-cross steers was amplified using gene-specific primers targeting the RNF34 and RNF128 genes. The PCR amplification products observed (intron 1) corresponded to the expected target fragments with good specificity. The PCR products were directly analyzed by restriction enzyme digestion and sequencing reactions.
The decision to investigate the 3′ UTR SNP of RNF34, despite the intron 1 of RNF128 yielding good specificity, was based on the distinct functional roles these regions play in gene expression and regulation. The 3′ UTR (untranslated region) of a gene is critical for the regulation of mRNA stability, localization, and translation efficiency.
Although selected for the specificity of PCR amplification (intron 1 region in RNF128) or potential to influence gene expression at post-transcriptional level (3′ UTR, both in RNF34), these regions happen within highly polymorphic genes and hence detect only a minor portion of genetic diversity. This is a region that was selected due to previous analysis implicating SNPs in the 3′ UTR as likely having biological significance, which would be an appropriate candidate gene region for exploring genetic impacts on economically important phenotypes.
3.2 Restriction endonuclease analysis and sequencing different genotypes
The PCR amplified products of RNF34 and RNF128 were subjected to SNP detection, genotyping, moreover, the two polymorphisms in both genes observed upon DNA sequencing using Sango tools (Shanghai, China). A single-nucleotide polymorphism site at nucleotide 588 in the RNF34 gene is an essential mediator of transcriptional and post-translational regulation (Fig. 1). PCR products from 255 samples were digested with the Bpu1102 restriction enzyme and resolved on a 3 % agarose gel for genotyping identification. A single band indicated a homozygous AA genotype, two bands indicated a homozygous GG genotype, and three bands indicated a heterozygous AG genotype, as shown in Fig. 1. Among the 255 Simmental-cross steers, the allele frequency for G at the 3′ UTR-G588A polymorphism site is 0.5882, whereas the frequency for allele A is 0.4118. Multi-comparison statistical analyses showed no statistically significant difference (P>0.05) in genotype distributions (Table 3). Using fragment length for RNF128, the sequencing results indicated that there was an SNP of I1-2380C>T in the first intron region. Following Spe I restriction, the following frequencies were found for the 213 samples: TT genotype of 0.5305, TC genotype of 0.0657, CC genotype of 0.4038, T allele of 0.5634, and C allele of 0.4366.
Of the 255 samples, 213 were successfully genotyped and included in the statistical analysis. Approximately 40 samples were excluded due to issues such as poor DNA quality, insufficient quantity after extraction, or incomplete data, which prevented successful PCR amplification and sequencing. These exclusions were necessary to ensure the accuracy and reliability of the results presented.
3.3 Association analyses of RNF34 and RNF128 polymorphisms with carcass and meat quality traits
One-way analysis of variance (ANOVA) was employed to explore the link between RNF34 polymorphisms and carcass traits of selected animals. Statistical analyses revealed that RNF34 3′ UTR (c.+588G>A) had a significant association with the carcass and meat quality traits, including tare weight, kidney weight, testis weight, and fat color score (P<0.05), as shown in Table 4.
Table 4RNF34 3′ UTR (c.+588G>A) association with the carcass and meat quality traits.
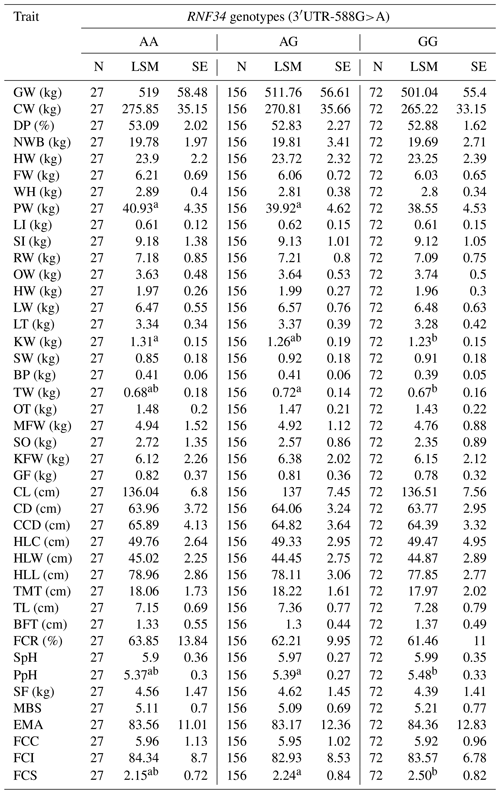
Values within a row that share the same letter (uppercase or lowercase) are not significantly different from each other (P > 0.05). Different letters indicate statistically significant differences (P < 0.05).
Table 5Association of RNF128 SNPs with carcass and meat quality traits in Simmental-cross steers.
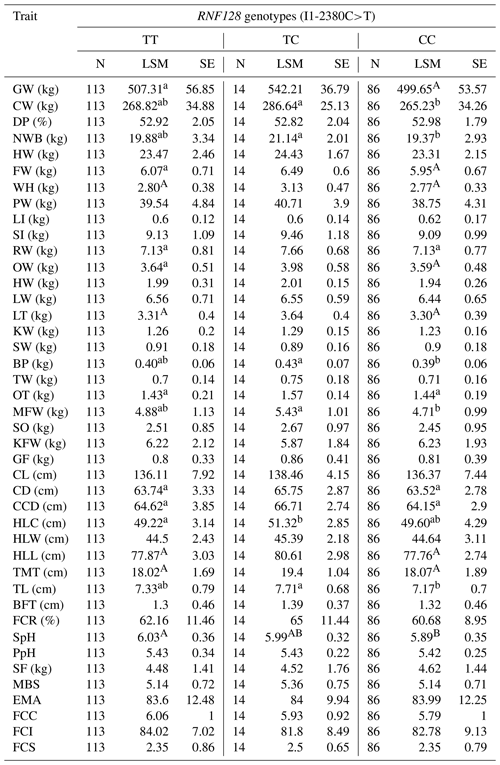
Values within a row that share the same letter (uppercase or lowercase) are not significantly different from each other (P > 0.05). Different letters indicate statistically significant differences (P <0.05). GW: gross weight; CW: carcass weight; DP: dressing percentage; NWB: net weight of bone; HW: head weight; FW: forepaw weight; WH: weight heels; LI: large intestine; SI: small intestine; RW: rumen weight; OW: omasum weight; HW: heart weight; LW: liver weight; LT: lung, trachea; KW: kidney weight; SW: spleen weight; BP: bull penis; TW: testes weight; OT: ox tail; MFW: mesenteric fat weight; SO: stomach oil; KFW: kidney fat weight; GF: genital fat; CL: carcass length; CD: carcass depth; CCD: carcass chest depth; HLC: hind leg circumference; HLW: hind leg width; HLL: hind leg length; TMT: thigh meat thickness; TL: thickness of loin; BFT: back fat thickness; FCR: fat coverage rate of carcass; SpH: slaughter pH; PpH: pH after acid exhausted; SF: shearing force; MBS: marbling score; EMA: eye muscle area; FCC: flesh color (color card); FCI: flesh (flesh-colored instrument); FCS: fat color score; LSM: least-squares mean; SE: standard error of mean.
One-way ANOVA and the least significant difference (LSD) test analyzed associations of the RNF128 gene polymorphisms with carcass traits to allow for multiple comparisons to be conducted concerning production traits. As illustrated in Table 5, the differences were significant between the different genotypes, involving forepaw weight, dressed weight, carcass depth, the thickness of waist flesh, carcass brisket depth, slaughter pH (P<0.05), lungs, trachea, and hind leg length (P<0.01).
Given the large number of characteristics analyzed, which includes correlated traits, there is potential for inflated type I error rates. To address this, a Bonferroni correction was applied to adjust the P values for multiple testing, ensuring that the reported significant associations are robust. Alternatively, the analysis was focused on a subset of essential characteristics most relevant to meat and carcass quality, reducing the potential for spurious findings.
Meat quality has great commercial importance for the animal husbandry industry. This industry heavily relies on the genetic background of the animals and their management, nutrition, and meat processing (Dong et al., 2019) although previous studies mainly focused on RNF34 and RNF128 were associated with cell differentiation and apoptosis (Verdin, 2010).
The observed differences in meat pH associated with the RNF34 and RNF128 SNPs may have important implications for meat quality. A lower pH in the animals with RNF34 3′ UTR-588. An allele may suggest a more rapid post mortem glycolysis, resulting in a faster decrease in its pH. This in turn can impact meat color, tenderness, and shelf life, which are salient factors for consumer acceptability. On the other hand, although not significant, pH increased by a slight level with I1-2380T allele in RNF128 could indicate a reduced rate for glycolysis that is another aspect that leads to differences on meat texture and water-holding capacity (Huff-Lonergan and Lonergan, 2005). This work revealed the possible influence of RNF34 and RNF128 on post mortem muscle metabolism, which may provide some new insight into meat quality traits, but more investigation is needed for use in marker-assisted selection.
Results show that RNF34 and RNF128 polymorphisms significantly correlates with certain meat quality traits in the study. These associations may be explained by RNF34’s regulatory effects on mRNA stability influencing muscle and fat tissue development as well as its role in T-cell function through transcriptional regulation. However, the effects of RNF34 on post-transcriptional regulation indicate that it might regulate protein synthesis in muscle cells and thus exert its influence via medication with these mRNA modifications.
RNF34 transcripts are highly enriched in brilliant cresyl blue (BCB) oocytes, suggesting that RNF34 may be involved in oocyte apoptosis (Wraith et al., 2009; Petroski and Deshaies, 2005b). And RNF34 plays a crucial role in the regulation of NOD1, RNF34, NF-κB pathways, which supports the idea that RNF34 is a negative regulator of the NOD1 pathway through direct interaction and ubiquitination of NOD1 (Skog et al., 2004; van Wijk et al., 2009). RNF128, an E3 ubiquitin-protein ligase, is an important gatekeeper of T cell unresponsiveness and uses a special single transmembrane protein with a split-function motif (Gavali et al., 2021). It may have a prominent role in CD4 T cell functions, including its survival, activation, and differentiation, where GRAIL acts in a well-characterized manner as it negatively regulates the responsiveness of T cell receptors and production of cytokine (Lett et al., 2004; Lineberry et al., 2008). However, new important roles of RNF34 and RNF128 are found in the present research.
The significant associations observed between RNF34 and RNF128 polymorphisms and meat quality traits in this study are consistent with findings from previous QTL and GWAS studies, which have highlighted the potential importance of these genes in livestock production. Although RNF34 and RNF128 are not traditionally associated with meat quality traits, their involvement in broader biological processes such as protein turnover, energy metabolism, and immune response provides a plausible link to the observed phenotypic variations (Petroski and Deshaies, 2005a; Schartner et al., 2009a). These results suggest that RNF34 and RNF128 may serve as novel markers for meat quality in cattle, warranting further investigation in larger and more genetically uniform populations to validate these findings.
In this study, SNPs of RNF34 and RNF128 are found to play important roles in meat quality traits and growth traits. SNP of RNF34 3′ UTR-588 G>A suggested that the AA genotype is significantly associated with testis weight and fat color score (Yu et al., 2017). The AG genotype is considerably associated with the weight of the kidney. Furthermore, the average production data for cattle of genotype AA are lower than those for cattle with AG or GG in pH after acid exhaustion and fat color score. The RNF128 gene I1-2380C>T shows that different genotypes significantly influence the carcass traits. There is a noticeable association between the TT genotype and the carcass weight, net weight of bone, bullwhip weight, mesenteric fat weight, and thickness of waist flesh. The TC genotype is significantly associated with slaughter pH. The CC genotype is significantly linked with hind leg circumference. The LSM of the carcass weight, net weight of bone, bullwhip weight, mesenteric fat weight, thickness of waist flesh, and hind leg circumference for the TC genotype higher than that of the TT or CC genotypes. The LSM of slaughter pH was calculated, and it is higher for the TT genotype than that for the TC or CC genotypes. So, we can select the excellent meat traits by genotypes of RNF34 and RNF128. Thus, this study supports the development of a novel theory about the cultivation of excellent beef using molecular biology techniques. Our results confirmed promising findings showing some previously mentioned associations but divergence from some studies, highlighting their limitations as further experimentations and required refinements for further validation of these associations.
This study provides evidence supporting the involvement of RNF34 and RNF128 in carcass and meat quality traits in Chinese Simmental-cross steers. The selection of these genes was informed by their known roles in ubiquitination and immune regulation as well as by QTL and GWAS studies that suggest a broader relevance to growth and meat quality traits in livestock. While the direct functions of these genes do not imply a causal link, their indirect effects on metabolic and cellular processes could influence economically important traits in cattle. Future research should aim to validate these findings in larger populations and explore the underlying mechanisms through which RNF34 and RNF128 may affect meat quality.
The data presented in this study are available from the corresponding authors upon reasonable request.
JS, AN, and JZZ: conceptualization. AN, JZZ, and SHW: methodology. QT and CYG: software and formal analysis. JZZ and AN: writing and preparation of original draft. SJ and YHJ: supervision and funding acquisition.
The contact author has declared that neither of the authors has any competing interests.
Animal experiments were conducted strictly with the highest ethical standards, adhering to the care and use of laboratory animals guidelines under the Jilin University Animal Care and Use Committee (permit no. SYXK (Ji) 2008-0010/0011).
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This work was supported by the Jilin Scientific and Technological Development Program (grant no. 20130522084JH). This study was funded by the National Natural Science Foundation of China (grant nos. 31660669 and 31372278).
This paper was edited by Henry Reyer and reviewed by Muhammad Kalim and one anonymous referee.
Afonso, J., Fortes, M. R. S., Reverter, A., Diniz, W., Cesar, A. S. M., Lima, A. O., Petrini, J., de Souza, M. M., Coutinho, L. L., Mourão, G. B., Zerlotini, A., Gromboni, C. F., Nogueira, A. R. A., and Regitano, L. C. A.: Genetic regulators of mineral amount in Nelore cattle muscle predicted by a new co-expression and regulatory impact factor approach, Sci. Rep., 10, 8436, https://doi.org/10.1038/s41598-020-65454-7, 2020.
Aly, S. M. and Sabri, D. M.: Next generation sequencing (NGS): a golden tool in forensic toolkit, Arch. Med. Sadowej Kryminol., 65, 260–271, https://doi.org/10.5114/amsik.2015.61029, 2015.
Anandasabapathy, N., Ford, G. S., Bloom, D., Holness, C., Paragas, V., Seroogy, C., Skrenta, H., Hollenhorst, M., Fathman, C. G., and Soares, L.: GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells, Immunity, 18, 535–547, https://doi.org/10.1016/s1074-7613(03)00084-0, 2003.
Antonaros, F., Olivucci, G., Cicchini, E., Ramacieri, G., Pelleri, M. C., Vitale, L., Strippoli, P., Locatelli, C., Cocchi, G., and Piovesan, A.: MTHFR C677T polymorphism analysis: A simple, effective restriction enzyme-based method improving previous protocols, Molecular Genetics & Genomic Medicine, 7, e628, https://doi.org/10.1002/mgg3.628, 2019a.
Antonaros, F., Olivucci, G., Cicchini, E., Ramacieri, G., Pelleri, M. C., Vitale, L., Strippoli, P., Locatelli, C., Cocchi, G., Piovesan, A., and Caracausi, M.: MTHFR C677T polymorphism analysis: A simple, effective restriction enzyme-based method improving previous protocols, Mol. Genet. Genomic. Med., 7, e628, https://doi.org/10.1002/mgg3.628, 2019b.
Bai, X. S., Zhang, C., Peng, R., Jiang, G. Q., Jin, S. J., Wang, Q., Ke, A. W., and Bai, D. S.: RNF128 Promotes Malignant Behaviors via EGFR/MEK/ERK Pathway in Hepatocellular Carcinoma, Onco. Targets Ther., 13, 10129–10141, https://doi.org/10.2147/ott.S269606, 2020.
Cai, C., Tang, Y.-D., Zhai, J., and Zheng, C.: The RING finger protein family in health and disease, Signal Transduction and Targeted Therapy, 7, 300, https://doi.org/10.1038/s41392-022-01152-2, 2022a.
Cai, C., Tang, Y. D., Zhai, J., and Zheng, C.: The RING finger protein family in health and disease, Signal Transduct. Target Ther., 7, 300, https://doi.org/10.1038/s41392-022-01152-2, 2022b.
Das, D. N., Karuthadurai, T., and Gnanasekaran, S.: Chapter 10 - Genomic selection: a molecular tool for genetic improvement in livestock, in: Advances in Animal Genomics, edited by: Mondal, S. and Singh, R. L., Academic Press, 141–163, https://doi.org/10.1016/B978-0-12-820595-2.00010-2, 2021.
Deng, T., Liang, M., Du, L., Li, K., Li, J., Qian, L., Xue, Q., Qiu, S., Xu, L., and Zhang, L.: Transcriptome Analysis of Compensatory Growth and Meat Quality Alteration after Varied Restricted Feeding Conditions in Beef Cattle, Int. J. Mol. Sci., 25, 2704, https://doi.org/10.3390/ijms25052704, 2024.
Deng, T., Wu, J., Abdel-Shafy, H., Wang, X., Lv, H., Shaukat, A., Zhou, X., Zhou, Y., Sun, H., Wei, P., Sun, N., Huang, Q., Xu, L., Liu, M., Lin, Y., Yang, L., and Hua, G.: Comparative Genomic Analysis of the Thiolase Family and Functional Characterization of the Acetyl-Coenzyme A Acyltransferase-1 Gene for Milk Biosynthesis and Production of Buffalo and Cattle, J. Agr. Food Chem., 71, 3325–3337, https://doi.org/10.1021/acs.jafc.2c07763, 2023.
Dong, C., Zhang, X., Liu, K., Li, B., Chao, Z., Jiang, A., Li, R., Li, P., Liu, H., and Wu, W.: Comprehensive Analysis of Porcine Prox1 Gene and Its Relationship with Meat Quality Traits, Animals, 9, 744, https://doi.org/10.3390/ani9100744, 2019.
Gavali, S., Liu, J., Li, X., and Paolino, M.: Ubiquitination in T-Cell Activation and Checkpoint Inhibition: New Avenues for Targeted Cancer Immunotherapy, Int. J. Mol. Sci., 22, 10800, https://doi.org/10.3390/ijms221910800, 2021.
Guo, P., Zhao, Z., Yan, S., Li, J., Xiao, H., Yang, D., Zhao, Y., Jiang, P., and Yang, R.: PSAP gene variants and haplotypes reveal significant effects on carcass and meat quality traits in Chinese Simmental-cross cattle, Arch. Anim. Breed., 59, 461–468, https://doi.org/10.5194/aab-59-461-2016, 2016.
Hashim, H. and Al-Shuhaib, M. B.: Exploring the Potential and Limitations of PCR-RFLP and PCR-SSCP for SNP Detection: A Review, Journal of Applied Biotechnology Reports, 6, 137–144, https://doi.org/10.29252/JABR.06.04.02, 2019.
Heissmeyer, V. and Rao, A.: E3 ligases in T cell anergy–turning immune responses into tolerance, Sci. STKE, 2004, pe29, https://doi.org/10.1126/stke.2412004pe29, 2004.
Huff-Lonergan, E. and Lonergan, S. M.: Mechanisms of water-holding capacity of meat: The role of postmortem biochemical and structural changes, Meat Sci., 71, 194–204, 2005.
Janowski, D., Salilew-Wondim, D., Torner, H., Tesfaye, D., Ghanem, N., Tomek, W., El-Sayed, A., Schellander, K., and Hölker, M.: Incidence of apoptosis and transcript abundance in bovine follicular cells is associated with the quality of the enclosed oocyte, Theriogenology, 78, 656–669.e5, https://doi.org/10.1016/j.theriogenology.2012.03.012, 2012.
Konica Minolta Sensing, Inc.: Precise Color Communication: Color Control from Perception to Instrumentation. Konica Minolta Sensing, Inc., https://www.konicaminolta.com/instruments/knowledge/color/pdf/color_communication.pdf (last access: 27 April 2025), 1998.
Korkmaz-Ağaoğlu, Ö., Elmaz, Ö., Akyüz, B., Zeytünlü, E., and Saatci, M.: Identifying polymorphism in some genes and their effects on growth performance in Honamli and hair goat breeds, Genetika, 51, 995–1008, https://doi.org/10.2298/GENSR1903995K, 2019.
Leal-Gutiérrez, J. D., Rezende, F. M., Elzo, M. A., Johnson, D., Peñagaricano, F., and Mateescu, R. G.: Structural equation modeling and whole-genome scans uncover chromosome regions and enriched pathways for carcass and meat quality in beef, Frontiers in Genetics, 9, 532, https://doi.org/10.3389/fgene.2018.00532, 2018.
Lett, D., Hsing, M., and Pio, F.: Interaction profile-based protein classification of death domain, BMC Bioinformatics, 5, 75, https://doi.org/10.1186/1471-2105-5-75, 2004.
Li, A., Qin, Q., Xiao, H., Fang, X., Jiang, P., Zhao, Z., and Yang, R.: Association of two sumo3 gene single nucleotide polymorphisms with carcass and meat quality traits of the chinese simmental cattle, J. Anim. Plant Sci., 27, 756–762, 2017.
Lineberry, N., Su, L., Soares, L., and Fathman, C. G.: The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane, J. Biol. Chem., 283, 28497–28505, https://doi.org/10.1074/jbc.M805092200, 2008.
Liu, L., Li, Y., Chen, G., and Chen, Q.: Crosstalk between mitochondrial biogenesis and mitophagy to maintain mitochondrial homeostasis, J. Biomed. Sci., 30, 86, https://doi.org/10.1186/s12929-023-00975-7, 2023.
Liu, Q. Y., Lei, J. X., Sikorska, M., and Liu, R.: A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer's patients and targets ATP6V0C for degradation, Mol. Neurodegener., 3, 1–16, 2008a.
Liu, Q. Y., Lei, J. X., Sikorska, M., and Liu, R.: A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer's patients and targets ATP6V0C for degradation, Mol. Neurodegener., 3, 4, https://doi.org/10.1186/1750-1326-3-4, 2008b.
Nawaz, A., Zhang, J., Meng, Y., Sun, L., Zhou, H., Geng, C., Liu, H., Jin, Y., and Ji, S.: Fatty acid profiles unveiled: gene expression in Yanbian yellow cattle adipose tissues offers new insights into lipid metabolism, Arch. Anim. Breed., 67, 469–480, https://doi.org/10.5194/aab-67-469-2024, 2024.
Nurieva, R. I., Zheng, S., Jin, W., Chung, Y., Zhang, Y., Martinez, G. J., Reynolds, J. M., Wang, S. L., Lin, X., Sun, S. C., Lozano, G., and Dong, C.: The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation, Immunity, 32, 670–680, https://doi.org/10.1016/j.immuni.2010.05.002, 2010.
Petroski, M. D. and Deshaies, R. J.: Function and regulation of cullin–RING ubiquitin ligases, Nat. Rev. Mol. Cell Biol., 6, 9–20, 2005a.
Petroski, M. D. and Deshaies, R. J.: Function and regulation of cullin-RING ubiquitin ligases, Nat. Rev. Mol. Cell Biol., 6, 9–20, https://doi.org/10.1038/nrm1547, 2005b.
Qi, Z. and Ding, S.: Transcriptional Regulation by Nuclear Corepressors and PGC-1α: Implications for Mitochondrial Quality Control and Insulin Sensitivity, PPAR Res., 2012, 348245, https://doi.org/10.1155/2012/348245, 2012.
Reshma, R. S. and Das, D. N.: Chapter 9 - Molecular markers and its application in animal breeding, in: Advances in Animal Genomics, edited by: Mondal, S. and Singh, R. L., Academic Press, 123–140, https://doi.org/10.1016/B978-0-12-820595-2.00009-6, 2021.
Sanglard, L. P., Snelling, W. M., Kuehn, L. A., Thallman, R. M., Freetly, H. C., Wheeler, T. L., Shackelford, S. D., King, D. A., and Spangler, M. L.: Genetic and phenotypic associations of mitochondrial DNA copy number, SNP, and haplogroups with growth and carcass traits in beef cattle, J. Anim. Sci., 101, skac415, https://doi.org/10.1093/jas/skac415, 2023a.
Sanglard, L. P., Snelling, W. M., Kuehn, L. A., Thallman, R. M., Freetly, H. C., Wheeler, T. L., Shackelford, S. D., King, D. A., and Spangler, M. L.: Genetic and phenotypic associations of mitochondrial DNA copy number, SNP, and haplogroups with growth and carcass traits in beef cattle, J. Anim. Sci., 101, skac415, https://doi.org/10.1093/jas/skac415, 2023b.
Schartner, J. M., Simonson, W. T., Wernimont, S. A., Nettenstrom, L. M., Huttenlocher, A., and Seroogy, C. M.: Gene related to anergy in lymphocytes (GRAIL) expression in CD4+ T cells impairs actin cytoskeletal organization during T cell/antigen-presenting cell interactions, J. Biol. Chem., 284, 34674–34681, 2009a.
Schartner, J. M., Simonson, W. T., Wernimont, S. A., Nettenstrom, L. M., Huttenlocher, A., and Seroogy, C. M.: Gene related to anergy in lymphocytes (GRAIL) expression in CD4+ T cells impairs actin cytoskeletal organization during T cell/antigen-presenting cell interactions, J. Biol. Chem., 284, 34674–34681, https://doi.org/10.1074/jbc.M109.024497, 2009b.
Sheng, X., Xia, Z., Yang, H., and Hu, R.: The ubiquitin codes in cellular stress responses, Protein Cell, 15, 157–190, https://doi.org/10.1093/procel/pwad045, 2023.
Skog, S., He, Q., Khoshnoud, R., Fornander, T., and Rutqvist, L. E.: Genes related to growth regulation, DNA repair and apoptosis in an oestrogen receptor-negative (MDA-231) versus an oestrogen receptor-positive (MCF-7) breast tumour cell line, Tumour Biol., 25, 41–47, https://doi.org/10.1159/000077722, 2004.
Tian, J., Zhao, Z., Zhang, L., Zhang, Q., Yu, Z., Li, J., and Yang, R.: Association of the leptin gene E2-169T>C and E3-299T>A mutations with carcass and meat quality traits of the Chinese Simmental-cross steers, Gene, 518, 443–448, https://doi.org/10.1016/j.gene.2012.11.071, 2013.
van Wijk, S. J. L., de Vries, S. J., Kemmeren, P., Huang, A., Boelens, R., Bonvin, A. M. J. J., and Timmers, H. T. M.: A comprehensive framework of E2–RING E3 interactions of the human ubiquitin–proteasome system, Mol. Syst. Biol., 5, 295, https://doi.org/10.1038/msb.2009.55, 2009.
Verdin, H.: Identification of novel disease gene for primary congenital glaucoma (PCG) through homozygosity mapping and next-generation sequencing strategies in a large consanguineous pedigree, Acta Ophthalmol., 88, Abstract 2163, https://doi.org/10.1111/j.1755-3768.2010.2163.x, 2010.
Wang, F. and Deng, X. W.: Plant ubiquitin-proteasome pathway and its role in gibberellin signaling, Cell Res., 21, 1286–1294, 2011a.
Wang, F. and Deng, X. W.: Plant ubiquitin-proteasome pathway and its role in gibberellin signaling, Cell Res., 21, 1286–1294, https://doi.org/10.1038/cr.2011.118, 2011b.
Wei, P., Pan, D., Mao, C., and Wang, Y. X.: RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1α and modulates brown fat cell metabolism, Mol. Cell Biol., 32, 266–275, https://doi.org/10.1128/mcb.05674-11, 2012.
Wraith, J. E., Baumgartner, M. R., Bembi, B., Covanis, A., Levade, T., Mengel, E., Pineda, M., Sedel, F., Topçu, M., Vanier, M. T., Widner, H., Wijburg, F. A., and Patterson, M. C.: Recommendations on the diagnosis and management of Niemann-Pick disease type C, Mol. Genet. Metab., 98, 152–165, https://doi.org/10.1016/j.ymgme.2009.06.008, 2009.
Xiao, H., Zhao, Z., Fang, X., Yu, H., Long, X., Jiang, P., and Yang, R.: Association of the ACSL5 gene G.33185918G>A and G.33186348C>T mutations with carcass and meat quality traits of Chinese Simmental-cross steers, J. Anim. Plant Sci., 26, 658–664 https://www.thejaps.org.pk/docs/v-26-03/14.pdf (last access: 27 April 2025), 2016.
Ye, T., Shaukat, A., Yang, L., Chen, C., Zhou, Y., and Yang, L.: Evolutionary and Association Analysis of Buffalo FABP Family Genes Reveal Their Potential Role in Milk Performance, Genes-Basel, 13, 600, https://doi.org/10.3390/genes13040600, 2022.
Yu, H., Zhao, Z., Yu, X., Li, J., Lu, C., and Yang, R.: Bovine lipid metabolism related gene GPAM: Molecular characterization, function identification, and association analysis with fat deposition traits, Gene, 609, 9–18, https://doi.org/10.1016/j.gene.2017.01.031, 2017.






