the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Investigating InDels in YAP and TAZ genes and their impact on growth characteristics in goats
Wanxia Zhao
Ziteng Wang
Yichen Lei
Xiaoqin Tang
Xiaohua Yi
Junyi Jiang
Jiapeng Li
Shuhui Wang
Xiuzhu Sun
Yes-associated protein (YAP) and a transcriptional co-activator with PDZ-binding motif (TAZ) genes are crucial for regulating the size of mammalian tissues and organs as well as for many biological processes such as bone formation, cell lineage determination, tissue regeneration, and cell proliferation. The purpose of this study was to characterize the YAP and TAZ gene polymorphisms in 266 Guanzhong Dairy Goats and 299 Shanbei White Cashmere Goats and to explore their potential relationship with growth characteristics such as body weight and body length. After genotyping and using PCR amplification and Sanger sequencing to find polymorphisms in the YAP and TAZ genes, five InDels loci were found in the goat YAP gene and three InDels loci in the TAZ gene. The findings of the association analysis demonstrated that the goats' body weight, height, cannon circumference, chest depth, chest breadth, and chest circumference were all substantially influenced by five InDels loci in the YAP gene (p<0.05). Goat body height, trunk breadth, trunk length, body length, and body weight were all substantially impacted by three InDels loci in the TAZ gene (p<0.05). In conclusion, eight InDels loci of goat YAP and TAZ were found in this study, and their impacts on goat phenotype were disclosed. These results might offer fresh avenues for boosting goat molecular breeding.
- Article
(1535 KB) - Full-text XML
- BibTeX
- EndNote
The transcriptional co-activator with a PDZ-binding motif (TAZ) and a yes-associated protein (YAP) are downstream effectors of the Hippo signaling pathway. TAZ and YAP have a 46 % amino acid sequence identity in mammals, and they can interact with transcription factors that contain the PPXY motif via the WW structural domain (Pocaterra et al., 2020). YAP and TAZ have garnered significant interest in recent times due to their vital functions in the formation and maintenance of tissue homeostasis in organisms. The activity of YAP and TAZ is essential for tissue-specific progenitor cell growth and cell proliferation during tissue renewal and regeneration (Heng et al., 2021; Piccolo et al., 2014). YAP and TAZ are involved in signaling during osteogenesis and play a critical role in the formation of bone and cartilage by modulating regulatory factors such as Runx2, Osx, and Sox9. The YAP and TAZ genes were knocked down using Prx1-Creo and Ose-Cre (Zarka et al., 2022; Xiong et al., 2018). As a result, mice experienced miscarriages during the perinatal stage, and the embryos displayed thoracic bleeding and skeletal abnormalities. Following TAZ gene knockdown using Col2-Cre, Y. Li et al. (2021) observed that young mouse pups showed growth retardation and underdevelopment of the sternum, ribs, and skull. In addition, it has been shown that the YAP gene plays an important role in muscle development in sheep and New Zealand white rabbits, regulating related genes such as MSTN, MYH6, and MYOD and other related genes associated with growth traits in livestock and poultry (Liu et al., 2023). Moreover, the YAP gene is an important pathway associated with susceptibility to brucellosis in sheep as well as a related candidate gene. As a result, YAP and TAZ contribute to bone production and are essential for animals' appropriate growth and development (X. Li et al., 2021).
According to Matin and Nadeau (2001), growth traits are quantitative traits regulated by microevolution polygenes that are impacted by both heredity and environment. Furthermore, more precise and efficient procedures are required for the selection of growth features. Animal breeding frequently employs molecular marker-assisted selection methods (Yan et al., 2014; Lin et al., 2021). One of the primary causes of molecular diversity among them is insertion/deletion. InDels are commonly employed in the research of the growth and economic aspects of domestic animals. It is essential in both genetic diversity and phenotypic differentiation (Cui et al., 2018). In light of the critical role that YAP/TAZ plays in growth and development, we examined the InDels of the YAP/TAZ gene in Guanzhong Dairy Goat and Shanbei White Cashmere Goat, assessing their effects on goat growth and development in order to identify key marker genes and furnish fundamental information that will expedite goat molecular breeding.
2.1 Sample collection and data collection
A total of 299 Shanbei White Cashmere Goats (SWCGs, Fugu, Shaanxi) and 266 Guanzhong Dairy Goats (GZDGs, Fuping, Shaanxi) were among the 565 local goats, ages 1–4 years, who were randomly chosen. Throughout the whole experiment, the management and feeding circumstances remained constant. All the goats' venous blood was drawn, and the growth characteristics were noted. The primary data consisted of the following: body height (BH), height at hip cross (HHC), body length (BL), rump length (RL), rump height (RH), rump width (RW), trunk length (TL), waist angle width (WAW), chest width (ChW), chest depth (ChD), chest circumference (ChC), cannon circumference (CaC), and body weight (BW). A single responsible individual measured each growth attribute to reduce measurement errors.
2.2 Genomic DNA extraction and DNA mixing pool construction
A Nanodrop1000 Spectrophotometer (Thermo Science, Waltham, MA, USA) was used to measure the concentration and purity (A260/280) of the extracted genomic DNA. The whole blood genomic DNA fast-extraction kit was kept at −80 °C. After diluting each sample to 20 ng µL−1 with ddH2O, it was kept at −20 °C. To identify putative InDels loci in the goat YAP/TAZ gene, 30 DNA samples were chosen at random from each breed and combined to create two DNA pools.
2.3 InDels loci discovery and primer design
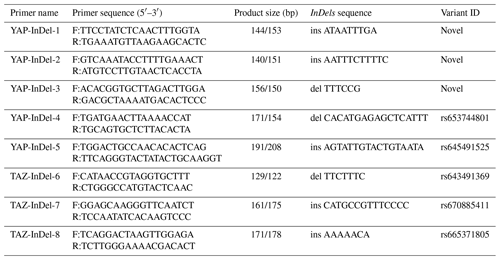
Five potential InDels loci were found in the goat YAP (GenBank accession no. NC_030822.1) gene, and three potential InDels loci were found in the goat TAZ (GenBank accession no. NC_030808.1) gene, according to the Ensemble database (http://asia.ensembl.org/index.html, last access: 20 May 2022) and the GGVD database (http://animal.nwsuaf.edu.cn/code/index.php/GoatVar, last access: 25 July 2022) (Fu et al., 2021). Primer Premier Software 5.0 was used to design the primers (Table 1).
2.4 Genotyping and DNA sequencing
The goat YAP/TAZ gene's InDels polymorphism was genotyped after the target gene was amplified using touch-down PCR, and the amplified products were separated by 3 % agarose gel electrophoresis. Generally speaking, genotypes were classified as homozygous (insertion/deletion: II), heterozygous (insertion/deletion: ID), and homozygous (deletion/deletion: DD). The YAP/TAZ gene mutation was confirmed by sequencing the PCR results of various genotypes.
2.5 Statistical analysis
Using the Botstein et al. (1980) approach, the genotype and allele frequencies of InDels mutations in goat breeds were determined. Using Haploview 4.2 software, linkage disequilibrium analysis of haplotypes at the InDels locus of the YAP/TAZ gene was carried out concurrently in two local breeds of sheep (Barrett, 2009). Utilizing Nei's (Nei and Roychoudhury, 1974; Nei and Li, 1979) methodology, the population's genetic diversity index was determined. The population's genetic variation was measured using the homozygosity (Ho), heterozygosity (He), and polymorphism information content (PIC), which served as an index of population polymorphism.
The statistical linear model is ijk, where Yijk is the observation of growth-related traits, μ is the overall mean for each trait, Ai is the effect of the ith age, Bj is the effect of the ith sex, Gk is the fixed effect of the genotype or combined genotype, and Eijk is a random residual. The fixed effect of genotypes was the main reason for the difference in growth traits. When the p value was less than 0.05, the difference was considered significant. Using GraphPad Prism 9 (GraphPad Software, USA) software, the Hardy–Weinberg equilibrium (HWE) state was analyzed using a chi-squared test (χ2 test). The correlation between the YAP/TAZ gene and growth traits of goats was analyzed by one-way ANOVA using SPSS 24 (IBM, USA) software.
3.1 Identification of genetic variants in the goats' YAP/TAZ gene
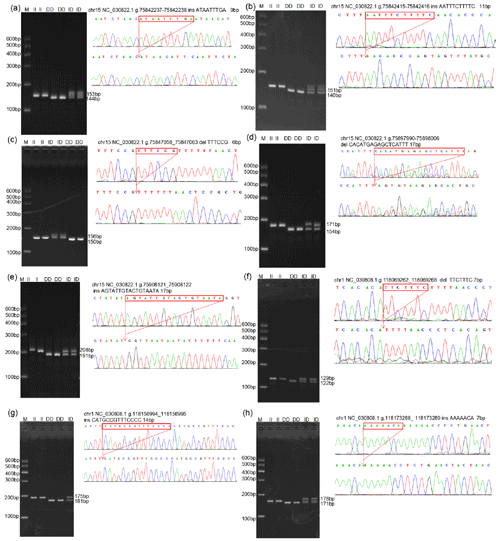
Through the analysis of DNA sequencing data, three InDels mutation sites (InDel-1 to InDel-8) were found in the TAZ gene's intron region, and five InDels mutation sites (InDel-1 to InDel-8) were found in the YAP gene's intron region (Fig. 1). By using 3 % agarose gel electrophoresis and sequencing, the genotyping of the InDels mutation sites of the YAP/TAZ gene was demonstrated (Fig. 2). Shanbei White Cashmere Goat is the only one with the following InDels loci: InDel-3 (NC_030822.1 g.75847058_75847063 del TTTCCG), InDel-4 (rs653744801, del CACATGAGAGCTCATTT), InDel-5 (rs645491525, ins AGTATTGTACTGTAATA), and InDel-6 (rs643491369, del TTCTTTC). Guanzhong Dairy Goat is the only one with InDel-2 (NC_030822.1 g.75842415-75842416 ins AATTTCTTTTC) and InDel-7 (rs670885411, ins CATGCCGTTTCCCC) (Fig. 2b, g). Both goat breeds have the InDel-1 (NC_030822.1 g.75842237-75842238 ins ATAATTTGA) and InDel-8 (rs665371805, ins AAAAACA) loci (Fig. 2a, h). It is important to note that InDel-1, InDel-2, and InDel-3 are unique in the GGVD database but not in the Ensemble database.
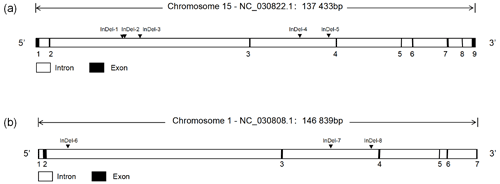
Figure 1The structure of the goat YAP/TAZ gene and the location of InDels mutation. (a) YAP gene structure map and InDels locus. (b) TAZ gene structure map and InDels locus.
3.2 Analysis of the genetic polymorphisms
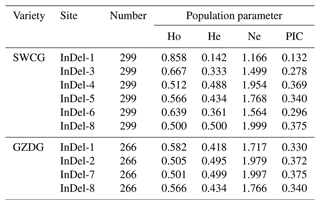
The genotype frequencies, allele frequencies, and polymorphism data of eight InDels loci of the YAP/TAZ gene were computed and examined in order to comprehend the variation of the YAP/TAZ gene in goats. It was discovered that the dominant genotypes in InDel-1, InDel-3, and InDel-6 were type II. In InDel-4 and InDel-8 it was ID, and in InDel-5 it was DD. In Shanbei White Cashmere Goat, the I allele frequencies of five InDels loci varied from 0.319 to 0.923, whereas the D allele frequencies varied from 0.077 to 0.681 (Table 2). The dominant genotype of InDel-1 and InDel-2 in Guanzhong Dairy Goat was the DD genotype, the dominant allele of InDel-7 was the ID genotype, and the dominant genotype of InDel-8 was the II genotype. Table 2 shows that the D allele frequencies of the four InDels loci ranged from 0.318 to 0.703, whereas the I allele rates fell between 0.297 and 0.682. The two goats had a genetic heterozygosity of 0.142 to 0.500 and an effective allele count of 1.166 to 1.999 (Table 3). The remaining InDels loci are in the moderate polymorphism range (0.25< PIC <0.5), with the exception of the Shanbei White Cashmere Goat InDel-1, which is in the low polymorphism range (PIC <0.25) (Table 3). Furthermore, the Hardy–Weinberg equilibrium of Guanzhong Dairy Goat and Shanbei White Cashmere Goat was confirmed using the χ2 test, and all of them were in equilibrium (p>0.05) (Table 2).
Table 2Genotype and gene frequencies of the InDels locus in the YAP/TAZ gene.
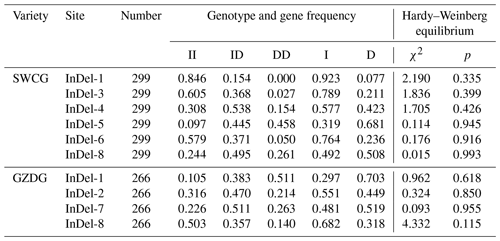
SWCG: Shanbei White Cashmere Goat; GZDG: Guanzhong Dairy Goat; II: insertion/insertion; ID: insertion/deletion; DD: deletion/deletion; I: insertion; D: deletion; χ2: chi-squared test; p: probability.
3.3 Linkage disequilibrium analysis
The number in the grid was D′, which was increased by 100 for esthetic reasons. The redder the color of the grid, the stronger the degree of linkage between the two loci, according to linkage disequilibrium analysis performed using the Haploview 4.2 software. In the YAP gene of Shanbei White Cashmere Goat, there was greater interlocking between InDel-3 and InDel-4 and less interlocking between the other loci (Fig. 3a). In Shanbei White Cashmere Goat, there was less chaining between the TAZ gene's InDels sites (Fig. 3b). The degree of interlocking between the InDel-7 and InDel-8 loci of the TAZ gene in Guanzhong Dairy Goat decreases with increasing interlocking between the InDel-1 and InDel-2 loci of the YAP gene (Fig. 3c, d).
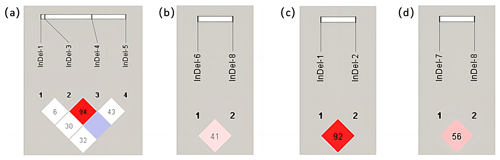
Figure 3Linkage disequilibrium analysis between different InDels loci of the YAP/TAZ gene in the two goats. (a) Linkage disequilibrium analysis between four InDels loci of the YAP gene in Shanbei White Cashmere Goat. (b) Linkage disequilibrium analysis between four InDels loci of the TAZ gene in Shanbei White Cashmere Goat. (c, d) Linkage disequilibrium analysis between the YAP and TAZ gene InDels loci in Guanzhong Dairy Goat.
3.4 Association analysis of genotype and growth traits
The results of the association analysis between the genotypes of various InDels loci and growth traits indicated that, for the growth trait of chest depth, genotype II of InDel-1 was considerably greater than genotype ID (p<0.01). Different genotypes in InDel-3 showed a substantial (p<0.05) correlation with both chest circumference and width. Chest width in InDel-4 was substantially associated (p<0.05) with different genotypes. Different genotypes in InDel-5 showed a substantial (p<0.05) correlation with body height, cross height, and circumference. Table 4 shows a substantial correlation (p<0.05) between different genotypes in InDel-6 and body height as well as a significant correlation (p<0.05) between different genotypes in InDel-8 and waist width.
Table 5 shows that there was a significant correlation between different genotypes of InDel-1, InDel-2, and InDel-7 with respect to body height, rump height, and body length (p<0.05); also, there was a significant correlation between different genotypes of InDel-8 and body weight (p<0.05) and between different genotypes of InDel-1 and rump width, cannon circumference, and body weight (p<0.01).
Surprisingly, across the two goats' various InDels loci, nearly all type-II homozygotes had the strongest growth performance (Tables 4 and 5). This showed that the TAZ genes InDel-7 and InDel-8 as well as the insertion mutations of the YAP genes InDel-1, InDel-2, and InDel-5 were advantageous. Furthermore, the Shanbei White Cashmere Goat YAP gene InDel-3 and InDel-4 DD type had the worst growth performance, whereas type-II homozygotes had the best growth performance. These findings suggested that InDel-3 and InDel-4 deletion mutations were detrimental variations. The data that are not significantly different are not provided, and Tables 4 and 5 mostly display the growth performance that differs from the growth features.
Table 4The effect of the InDels locus of the YAP/TAZ gene on growth traits of Shanbei White Cashmere Goat.
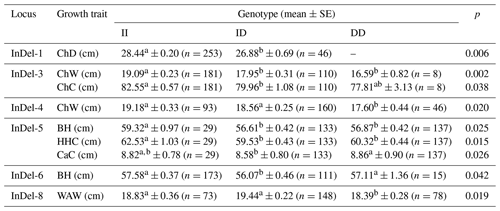
ChD: chest depth; ChW: chest width; ChC: chest circumference; BH: body height; HHC: height at hip cross; CaC: cannon circumference; WAW: waist angle width; SE: standard error; II: insertion/insertion; ID: insertion/deletion; DD: deletion/deletion. a,b Values within a row with different superscripts differ significantly at p<0.05.
Table 5Effects of the InDels locus of the YAP/TAZ gene on growth traits of Guanzhong Dairy Goat.
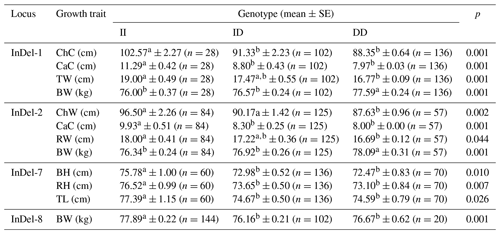
ChC: chest circumference; CaC: cannon circumference; TW: thurl width; BW: body weight; ChW: chest width; RW: rump width; BH: body height; RH: rump height; TL: trunk length; SE: standard error; II: insertion/insertion; ID: insertion/deletion; DD: deletion/deletion. a,b Values within a row with different superscripts differ significantly at p<0.05.
One of the main causes of genetic diversity is insertion and deletion mutation. Animal growth and development may be impacted by InDels mutations in the genome due to a variety of phenotypic and molecular effects (Pagel et al., 2019). In this study, three InDels were discovered in the TAZ gene's intron area, and five InDels were discovered in the YAP gene's intron in two local goat breeds. Table 2 shows that most polymorphism information content was moderately polymorphic, and the highest population heterozygosity (0.418–0.500) and effective allele count (1.717–1.999) were found in InDel-4, InDel-5, and InDel-8 of Shanbei White Cashmere Goat and InDel-1, InDel-7, and InDel-8 of Guanzhong Dairy Goat, respectively. These results suggest that the population has more evenly distributed alleles of these InDels loci, the highest degree of genetic variation, and a richer genetic diversity overall. Furthermore, the frequency of the D allele is low, and there is no DD genotype in InDel-1 of Shanbei White Cashmere Goat. Either too little sample size or homozygous embryo mortality could be the cause of this.
According to Crow (1988), the Hardy–Weinberg law is the basis of contemporary genetics and the basis of population genetics. HWE is able to determine the reliability of population survey data in addition to estimating gene frequency from phenotype and genotype frequency in terms of population genetics of human genetic markers. The deviation from the Hardy–Weinberg equilibrium happens when there are selection, drift, population mixing, or non-random mating forms (Kang and Shin, 2004; Wigginton et al., 2005). All of the InDels loci of the YAP/TAZ gene were in Hardy–Weinberg equilibrium, according to the study's χ2 test results (Table 2).
Numerous studies have suggested that variations in InDels may impact an animal's growth qualities, and InDels are frequently employed in molecular marker-assisted breeding as a molecular marker linked to significant economic attributes (Wu et al., 2020). For example, Wang et al. (2020) and Zhang et al. (2021) found a substantial correlation between litter size and the 21 bp InDel mutation in the ATBF1 gene and the 16 bp InDel mutation in the AKAP13 gene. Male sheep's tail fat deposition was considerably impacted by the FTO gene's InDels mutation (Wang et al., 2021). Chest depth in Lanzhou big-tailed sheep and body weight, body height, and other body-measuring features in Ruxi black-headed sheep were strongly impacted by the 7 bp InDel locus in the KDM3B gene (Kang et al., 2022). In addition, the Del/Del genotype of the rs669452874 locus of the GLIPR1L1 gene significantly affected the height and length of Dazu black goats, Hechuan white goats, and Inner Mongolian Cashmere goats (Saleh et al., 2023).
According to Shaul (2017), introns can boost mRNA translation efficiency and stability in the cytoplasm. Enhancers and silencers are examples of cis-acting regions in introns that influence mRNA shearing. Additionally, intron mutations may cause mRNAs to detect incorrect sequences during splicing, resulting in improper removal of introns, which may alter open reading frames (Sarkar, 2022). The YAP gene's introns containing the mutations of InDel-1, InDel-2, InDel-3, InDel-4, and InDel-5 in this study had a substantial impact on the goats' body height, cross height, cannon circumference, rump breadth, and body weight. The goats' body height, rump height, body length, and body weight were all positively impacted by the mutations of InDel-6, InDel-7, and InDel-8 in the TAZ gene's introns. Important osteogenesis mediators include TAZ and YAP. According to Zaidi et al. (2004), YAP can block the Runx2-dependent transcriptional activation of osteocalcin (OCN). Furthermore, osteogenesis is sped up by YAP deficiency and inhibited by high expression of YAP in bone marrow mesenchymal stem cells. According to studies, YAP prevents endochondral ossification during the growth and healing of bones. Defects in bone formation occur in vivo when the YAP/TAZ combination is absent (Yang et al., 2019). The Hippo signaling pathway plays an important role in the control of organ size in animals, and YAP is a transcriptional co-activator and a key effector in the Hippo signaling pathway. The results of Ishihara and Nishina (2018) showed that YAP/TAZ regulates organ size by modulating cellular tone: in addition, in a chimeric mouse model, hepatocytes with impaired YAP gene expression activity undergo apoptosis and are cleared from the liver. Therefore, YAP/TAZ has the function of controlling the number and quality of cells in the process of organogenesis, and the organ size is closely related to the body size of animals. Thus, YAP/TAZ affects the body size of animals by influencing the organ size. Situated at the core of an intricate signal network with multiple biological purposes, YAP/TAZ is crucial for controlling animal growth.
Briefly, to the best of our knowledge, there is a wide range of differences for body measurements and the body indices among breeds of goats (Saleh et al., 2022). The screening and function identification of InDel markers is beneficial for further developing and utilizing high-quality genes and germplasm resources.
This investigation identified YAP/TAZ polymorphisms from northern Shanbei White Cashmere Goat and Guanzhong Dairy Goat, determined the candidate InDels, and uncovered their association with phenotypic data of the productive performance. These InDels loci may provide a database for genetic analysis and molecular marker-assisted breeding in goat populations. The economic efficiency of the goat industry can be increased by applying these insights to the breeding process.
Animal data were not deposited in an official repository and are confidential.
WZ wrote the first draft, performed the genotyping experiment, collated and analyzed the data, and managed the data. ZW and YL were both involved in the genotyping experiments. XT was responsible for data analysis and article revision. XY participated in sample collection and article revision. JJ and JL were involved in the collection of samples and data. SW and XS proposed the idea of this study and participated in the collection of samples.
The contact author has declared that none of the authors has any competing interests.
In this study, all implemented experiments were approved by the Animal Use Review Committee of Northwest A&F University (NWAFU314020038) and fully complied with local animal welfare guidelines, laws, and policies.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
We are grateful to Fugu County Hara Debt Agricultural Development Co., Ltd., which provided the experimental samples. We are also grateful for the Ensemble genome database created by the European Institute of Bioinformatics, to the Wellcome Trust Sanger Institute, and for the GGVD database created by the Yu Jiang team at the College of Animal Science and Technology, Northwest A&F University, for providing us with whole-genome sequencing data.
This research has been supported by the Key Research and Development Program of Shaanxi Province (grant no. 2018ZDXM-NY-040).
This paper was edited by Henry Reyer and reviewed by Yun Ma, Ahmed A. Saleh, and one anonymous referee.
Barrett, J. C.: Haploview: Visualization and analysis of SNP genotype data, Cold Spring Harbor Protocols, 71, pdb.ip71, https://doi.org/10.1101/pdb.ip71, 2009.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, 1980.
Crow, J. F.: Eighty years ago: the beginnings of population genetics, Genetics, 119, 473–476, https://doi.org/10.1093/genetics/119.3.473, 1988.
Cui, Y., Yan, H., Wang, K., Xu, H., Zhang, X., Zhu, H., Liu, J., Qu, L., Lan, X., and Pan, C.: Insertion/Deletion Within the KDM6A Gene Is Significantly Associated With Litter Size in Goat, Front. Genet., 9, 91, https://doi.org/10.3389/fgene.2018.00091, 2018.
Fu, W., Wang, R., Yu, J., Hu, D., Cai, Y., Shao, J., and Jiang, Y.: GGVD: A goat genome variation database for tracking the dynamic evolutionary process of selective signatures and ancient introgressions, J. Genet. Genomics, 48, 248–256, https://doi.org/10.1016/j.jgg.2021.03.003, 2021.
Heng, B. C., Zhang, X., Aubel, D., Bai, Y., Li, X., Wei, Y., Fussenegger, M., and Deng, X.: An overview of signaling pathways regulating YAP/TAZ activity, Cell. Mol. Life Sci., 78, 497–512, https://doi.org/10.1007/s00018-020-03579-8, 2021.
Ishihara, E. and Nishina, H.: The Hippo-YAP Pathway Regulates 3D Organ Formation and Homeostasis, Cancers, 10, 122, https://doi.org/10.3390/cancers10040122, 2018.
Kang, S. H. and Shin, D.: The size of the chi-square test for the Hardy-Weinberg law, Hum. Hered., 58, 10–17, https://doi.org/10.1159/000081452, 2004.
Kang, Y., Bi, Y., Tang, Q., Xu, H., Lan, X., Zhang, Q., and Pan, C.: A 7-nt nucleotide sequence variant within the sheep KDM3B gene affects female reproduction traits, Anim. Biotechnol., 33, 1661–1667, https://doi.org/10.1080/10495398.2021.1929270, 2022.
Li, X., Wu, Q., Zhang, X., Li, C., Zhang, D., Li, G., Zhang, Y., Zhao, Y., Shi, Z., Wang, W., and Li, F.: Whole-Genome Resequencing to Study Brucellosis Susceptibility in Sheep, Front. Genet., 8, 653927, https://doi.org/10.3389/fgene.2021.653927, 2021.
Li, Y., Yang, S., Qin, L., and Yang, S.: TAZ is required for chondrogenesis and skeletal development, Cell Discovery, 7, 26, https://doi.org/10.1038/s41421-021-00254-5, 2021.
Lin, W., Ren, T., Li, W., Liu, M., He, D., Liang, S., Luo, W., and Zhang, X.: Novel 61-bp InDel of RIN2 Is Associated With Fat and Hatching Weight Traits in Chickens, Front. Genet., 12, 672888, https://doi.org/10.3389/fgene.2021.672888, 2021.
Liu, Y. Q., Chen, M. J., Wang, Y. J., Xu, H. F., Li, M., and Yu, G. Q.: YAP1 Regulating the Muscle Growth and Development in New ZealandWhite Rabbits, Chinese Journal of Biochemistry and Molecular Biology, 9, 1765–1775, 2023.
Matin, A. and Nadeau, J. H.: Sensitized polygenic trait analysis, Trends Genet., 17, 727–731, https://doi.org/10.1016/s0168-9525(01)02528-8, 2001.
Nei, M. and Li, W. H.: Mathematical model for studying genetic variation in terms of restriction endonucleases, P. Natl. Acad. Sci. USA., 76, 5269–5273, https://doi.org/10.1073/pnas.76.10.5269, 1979.
Nei, M. and Roychoudhury, A. K.: Sampling variances of heterozygosity and genetic distance, Genetics, 76, 379390, https://doi.org/10.1093/genetics/76.2.379, 1974.
Pagel, K. A., Antaki, D., Lian, A., Mort, M., Cooper, D. N., Sebat, J., Iakoucheva, L. M., Mooney, S. D., and Radivojac, P.: Pathogenicity and functional impact of non-frameshifting insertion/deletion variation in the human genome, PLoS Comput. Biol., 15, e1007112, https://doi.org/10.1371/journal.pcbi.1007112, 2019.
Piccolo, S., Dupont, S., and Cordenonsi, M.: The biology of YAP/TAZ: hippo signaling and beyond, Physiol. Rev., 94, 1287–1312, https://doi.org/10.1152/physrev.00005, 2014.
Pocaterra, A., Romani, P., and Dupont, S.: YAP/TAZ functions and their regulation at a glance, J. Cell. Sci., 133, jcs230425, https://doi.org/10.1242/jcs.230425, 2020.
Saleh, A. A., Rashad, A. M. A., Hassanine, N. N. A. M., Sharaby, M. A., and Zhao, Y.: Morphological body measurements, body indices, and their genetic background for several Chinese goat breeds, Trop. Anim. Health Pro., 54, 204, https://doi.org/10.1007/s11250-022-03203-w, 2022.
Saleh, A. A., Xue, L., and Zhao, Y.: Screening InDels from the whole genome to identify the candidates and their association with economic traits in several goat breeds, Funct. Integr. Genomic., 23, 58, https://doi.org/10.1007/s10142-023-00981-w, 2023.
Sarkar, A., Panati, K., and Narala, V. R.: Code inside the codon: The role of synonymous mutations in regulating splicing machinery and its impact on disease, Mutation research, Reviews in Mutation Research, 790, 108444, https://doi.org/10.1016/j.mrrev.2022.108444, 2022.
Shaul, O.: How introns enhance gene expression, Int. J. Biochem. Cell Biol., 91, 145–155, https://doi.org/10.1016/j.biocel, 2017.
Wang, K., Hui, Y., Zhang, S., Wang, M., Yan, H., Zhu, H., Qu, L., Lan, X., and Pan, C.: A deletion mutation within the ATBF1 gene is strongly associated with goat litter size, Anim. Biotechnol., 31, 174–180, https://doi.org/10.1080/10495398.2018.1561459, 2020.
Wang, S., Liu, S., Yuan, T., and Sun, X.: Genetic effects of FTO gene insertion/deletion (InDel) on fat-tail measurements and growth traits in Tong sheep, Anim. Biotechnol., 32, 229–239, https://doi.org/10.1080/10495398.2019.1680379, 2021.
Wigginton, J. E., Cutler, D. J., and Abecasis, G. R.: A note on exact tests of Hardy-Weinberg equilibrium, Am. J. Hum. Genet., 76, 887–893, https://doi.org/10.1086/429864, 2005.
Wu, M., Zhao, H., Tang, X., Li, Q., Yi, X., Liu, S., and Sun, X.: Novel InDels of GHR, GHRH, GHRHR and Their Association with Growth Traits in Seven Chinese Sheep Breeds, Animals, 10, 883, https://doi.org/10.3390/ani10101883, 2020.
Xiong, J., Almeida, M., and O'Brien, C. A.: The YAP/TAZ transcriptional co-activators have opposing effects at different stages of osteoblast differentiation, Bone, 112, 1–9, https://doi.org/10.1016/j.bone.2018.04.001, 2018.
Yan, Y., Yi, G., Sun, C., Qu, L., and Yang, N.: Genome-wide characterization of insertion and deletion variation in chicken using next generation sequencing, PloS one, 9, e104652, https://doi.org/10.1371/journal.pone.0104652, 2014.
Yang, B., Sun, H., Chen, P., Fan, N., Zhong, H., Liu, X., Wu, Y., and Wang, J.: YAP1 influences differentiation of osteoblastic MC3T3-E1 cells through the regulation of ID1, J. Cell Physiol., 234, 14007–14018, https://doi.org/10.1002/jcp.28088, 2019.
Zaidi, S. K., Sullivan, A. J., Medina, R., Ito, Y., van Wijnen, A. J., Stein, J. L., Lian, J. B., and Stein, G. S.: Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription, EMBO J., 23, 790–799, https://doi.org/10.1038/sj.emboj.7600073, 2004.
Zarka, M., Haÿ, E., and Cohen-Solal, M.: YAP/TAZ in Bone and Cartilage Biology, Front. Cell Dev. Biol., 9, 788773, https://doi.org/10.3389/fcell.2021.788773, 2022.
Zhang, X., Yuan, R., Bai, Y., Yang, Y., Song, X., Lan, X., and Pan, C.: A deletion mutation within the goat AKAP13 gene is significantly associated with litter size, Anim. Biotechnol., 34, 350–356, https://doi.org/10.1080/10495398.2021.1968418, 2021.