the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Inheritance of body size and ultrasound carcass traits in yearling Anatolian buffalo calves
Mustafa Tekerli
The body size and ultrasound carcass traits are related to the growth and muscling of animals. These characters promise future improvement through genetic selection in animal breeding. In breeding programs, knowing the (co)variance components serves to reveal the performance differences among animals and detection of suitable traits for selection. The research was carried out with 313 Anatolian buffalo calves born in 2019 at 36 farm operations. The least-square means for body weight (BW), wither height (WH), rump height (RH), body length (BL), chest width (CW), hip width (HW), chest circumference (CC), cannon-bone circumference (CBC), longissimus muscle area (LMA), longissimus muscle depth (LMD), and subcutaneous fat thickness (SFT) in yearling calves were 175.41 ± 2.06 kg, 108.35 ± 0.34, 111.85 ± 0.37, 103.74 ± 0.41, 33.93 ± 0.23, 30.56 ± 0.23, 135.18 ± 0.60, 15.69 ± 0.08 cm, 19.36 ± 0.45 cm2, 3.086 ± 0.028, and 0.655 ± 0.006 cm, respectively. The direct heritabilities for BW, WH, RH, BL, CW, HW, CC, CBC, LMA, LMD, and SFT were 0.334 ± 0.032, 0.483 ± 0.044, 0.473 ± 0.043, 0.441 ± 0.041, 0.364 ± 0.034, 0.432 ± 0.040, 0.435 ± 0.040, 0.226 ± 0.021, 0.0001 ± 0.000, 0.300 ± 0.026, and 0.539 ± 0.046, respectively. The genetic and phenotypic correlations predicted in this study ranged from 0.02 to 0.90. All the genetic and phenotypic correlations among body size and ultrasound carcass traits were significant (P<0.01), except for the genetic correlation between CW and HW. Some polymorphisms in PLAG1, NCAPG, LCORL, and HMGA2 genes were analyzed. Two single-nucleotide polymorphisms (SNPs) for PLAG1 and NCAPG genes were found to be monomorphic in this buffalo population. Meanwhile, the effects of two SNPs in the LCORL and HMGA2 genes were not significant but showed some tendencies in the aspects of least-square means. The results of the study indicated that the Anatolian buffaloes have the potential to improve in growth and muscling characteristics.
- Article
(472 KB) - Full-text XML
- BibTeX
- EndNote
The buffalo is a robust and thrifty animal. It is one of the species that people benefit from. e.g., meat, milk, leather, and manure production, and they are used as draw animal in some distinct regions of the world. When compared, water buffalo have fallen short cattle in the aspect of production, but it is still an alternative production resource throughout the world (İzmen and Spöttel, 1937; Uslu, 1970; Şahin et al., 2013; Koçak et al., 2019). Bubalus, known as water buffalo, is in the same subfamily with Bos, Syncerus, and Bison. The order of water buffalo in taxonomy is as follows: Mammalia class, Artiodactyla order, Bovidae family, Bovinae subfamily, Bubalus genus, and Bubalus bubalis species. Domestic buffaloes are divided into two subspecies. These are riverine (Bubalus bubalis bubalis, 2n=50 chromosomes) and swamp (Bubalus bubalis kerabau, 2n=48 chromosomes) buffaloes (Soysal, 2009; ITIS, https://itis.gov, last access: 14 November 2022).
Studies about body measurements are generally conducted on adult buffaloes (Sindhu and Pal, 1980; Peeva, 1991; Ahmad et al., 2013; Dhillod et al., 2017; de Melo et al., 2018; Riaz et al., 2018; Korejo et al., 2019; Dahiya et al., 2020; Mirza et al., 2020). However, research focusing on yearling buffaloes are scarce (Gürcan et al., 2011; Gavit et al., 2013; Çelikeloğlu et al., 2019; Ağyar et al., 2022). Some scientists (Thevamanoharan et al., 2001; Mirza et al., 2020) were interested in genetic parameters of lactating buffaloes except for Vankov and Peeva (1994). The genetic background and relationships of body size traits have been relatively uncertain in different breeds even today.
Ultrasound carcass traits in adult buffaloes were measured and reported by Harada et al. (1997), Jorge et al. (2007), Rebak et al. (2010), and Singh et al. (2018). Taborda et al. (2015) notified the community that the heritabilities for longissimus muscle area (LMA) and rump fat thickness were 0.256 and 0.214.
The number of studies aiming to understand the genetic basis of growth and development in humans and cattle showed a significant increase in the last decades. Single-nucleotide polymorphisms (SNPs) in PLAG1, NCAPG, LCORL, and HMGA2 genes were found to be associated with height in humans (Gudbjartsson et al., 2008). Pryce et al. (2011) also reported that the same genes had an effect on RH in cattle by similar functionalities.
The objectives of this study were to reveal the hereditary background of body size and ultrasound carcass traits and enlighten some SNPs in PLAG1, NCAPG, LCORL, and HMGA2 genes in yearling Anatolian buffaloes.
This study was reviewed and approved by the experimental animal ethics committee of Afyon Kocatepe University (AKUHADYEK, 49533702/258 and 210/20).
The data consisted of 2165 body measurements and 730 ultrasound carcass trait records from a total of 313 yearling Anatolian buffalo registered to the “Community Based Buffalo Improvement Project” governed by the General Directorate of Agricultural Research and Policies of the Ministry of Agriculture and Forestry. All measurements were taken from February to August 2019 in 36 farm operations.
Body size traits of an animal with a normal posture standing on a weighing scale were measured using measuring stick, caliper, and tape as described by Batu (1951), Kendir (1960), Arpacık (1982), and Gilbert et al. (1993). Body measurements were wither height (the highest point on the withers), rump height (the highest point on the rump), body length (the distance between caput humerii and tuber ischiadicum), chest width (the distance between two caput humerii), hip width (the distance between two Tuber coxae), chest circumference (circumference immediately posterior of the front legs), and cannon-bone circumference (the circumference of the left metacarpus at its narrowest).
After the body weight and measurements were recorded, the region between the 13th dorsal vertebra and the first lumbar vertebra (Hwang et al., 2014) on the left side of the animals was scanned for the ultrasound measurements. The hair of the measurement area was clipped, and then all the hairs were removed with the help of a razor blade to obtain a good-quality image. The conductive medium was ultrasound gel. The SIUI CTS-800 scanner with linear and back-fat probes was used for imaging. An instant ultrasound image was obtained for each animal. The images were processed by ImageJ (Schneider et al., 2012) software after the calibration of pixels (Fig. 1).
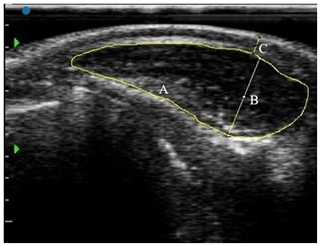
Figure 1The real-time ultrasound image and its interpretation in a buffalo calf (borders A and depth B of longissimus muscle, and subcutaneous fat thickness (SFT) labeled C).
The values at 12 months of age for all traits were estimated from successive measurements by interpolation or extrapolation as described by Gürtan (1979) using an in-house add-in running in Microsoft Excel.
Genomic DNA was extracted from blood samples using a modified method described by Boom et al. (1990). We selected SNPs coded as rs109231213 (Karim et al., 2011) and c.132T > G (Setoguchi et al., 2009) for PLAG1 and NCAPG genes. These SNPs were examined for nucleotide variation by the Sanger sequence (ABI 3500 genetic analyzer) using pool DNA (12 animals for each). AX-85166825 and AX-85179490 coded SNPs located in the upstream and downstream regions of LCORL and HMGA2 genes were preferred. The PCR-RFLP (restriction fragment length polymorphism) method was applied for the detection of variants. For this purpose, BccI and BmrI restriction enzymes were used. Genotyping was completed by agarose gel electrophoresis and imaging (Bio-Vision, Vilber Lourmat).
Table 1Least-square means for body size traits in Anatolian buffalo.
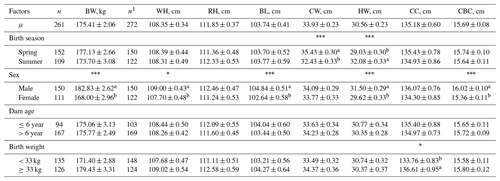
Abbreviations: BW, body weight; WH, wither height; RH, rump height; BL, body length; CW, chest width; HW, hip width; CC, chest circumference; CBC, cannon-bone circumference. 1 The number of animals used in the analysis for WH, RH, BL, CW, HW, CC, and CBC. * P<0.05; P<0.01; P<0.001. a,b Means with different superscripts within a variable differ significantly (P<0.05).
Table 2Least-square means for ultrasound carcass traits in Anatolian buffalo.
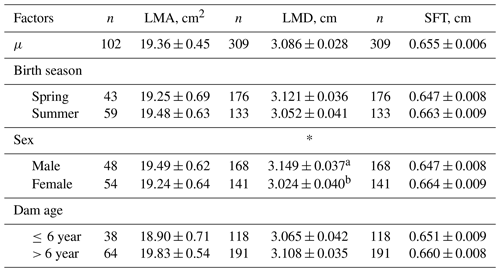
Abbreviations: LMA, longissimus muscle area; LMD, longissimus muscle depth; SFT, subcutaneous fat thickness. * P<0.05.
The effects of farm, birth season, sex, dam age and birth weight on body measurements and ultrasound carcass traits (except for birth weight) were analyzed by the method of least squares using the following model:
where Yijklmn=nth observation in the mth birth weight, lth dam age, kth sex, jth birth season, and ith farm; μ = the overall mean; Fi= the effect of ith farm (i = 1, 2, …, 35, and 36); BSj= the effect of jth birth season (j = spring, summer); Sk= the effect of kth sex (k = male, female); DAl= the effect of lth dam age (l , > 6 year); BWm= the effect of mth birth weight (m = < 33, ≥ 33 kg); eijklmn= random error N (0, σ2).
Table 3Estimation of variance components for body size and ultrasound carcass traits in Anatolian buffalo.
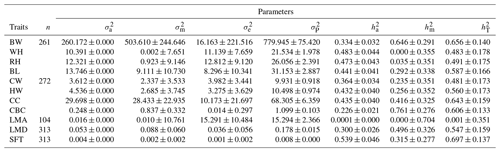
Abbreviations: BW, body weight; WH, wither height; RH, rump height; BL, body length; CW, chest width; HW, hip width; CC, chest circumference; CBC, cannon-bone circumference; LMA, longissimus muscle area; LMD, longissimus muscle depth; SFT, subcutaneous fat thickness; , direct additive genetic variance; , maternal genetic variance; , residual variance; , phenotypic variance; , direct additive heritability; , maternal heritability; , total heritability.
The farms with at least four animals for all traits were used in the analyses. The birth season consisted of spring and summer subgroups. Sex and dam age were divided into two groups (male and female, ≤ 6 and > 6 years of age). The least-square analyses were applied by Minitab 18, and the Tukey test was used for multiple comparisons.
Estimation of (co)variance components for body measurements and ultrasound carcass traits were obtained by restricted maximum likelihood (REML) using WOMBAT (Meyer, 2007) software, considering univariate animal model and numerator relationship matrix to obtain a more accurate breeding value (Meyer, 1992; Çinkaya et al., 2019). The model used for the analyses was as follows:
where Y is the vector of observations, β is the vector of nongenetic significant fixed effects, a is the vector of random additive genetic effects, m is the vector of random maternal genetic effects, and e is the unknown random vector of residuals. X, Za, and Zm are incidence matrices associating observations to fixed, additive genetic, and maternal genetic effects, respectively.
The genetic correlations for body measurements and ultrasound carcass traits were calculated from the estimated breeding values (EBVs) of related traits with the following formula (Calo et al., 1973; Falconer and Mackay, 1996; Mitchell et al., 2005);
where rG(XY) is the genetic correlation between X and Y traits, CovXY is the covariance between EBVs for X and Y, is the direct additive genetic variance of trait X, and is the direct additive genetic variance of trait Y.
Table 4Genetic (upper diagonal) and phenotypic (lower diagonal) correlations among body size and ultrasound carcass traits of Anatolian buffalo.
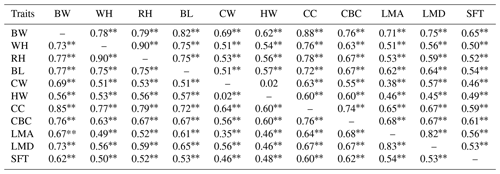
Abbreviations: BW, body weight; WH, wither height; RH, rump height; BL, body length; CW, chest width; HW, hip width; CC, chest circumference; CBC, cannon-bone circumference; LMA, longissimus muscle area; LMD, longissimus muscle depth; SFT, subcutaneous fat thickness. P<0.01.
Table 5Direct (diagonal) and correlated responses of selection for body size and ultrasound carcass traits in Anatolian buffalo.
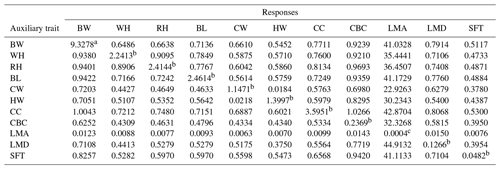
Abbreviations: BW, body weight; WH, wither height; RH, rump height; BL, body length; CW, chest width; HW, hip width; CC, chest circumference; CBC, cannon-bone circumference; LMA, longissimus muscle area; LMD, longissimus muscle depth; SFT, subcutaneous fat thickness. a: kg; b: cm; c: cm2.
The phenotypic correlations were calculated as Pearson correlation coefficient with the adjusted data for significant environmental effects. Corrections were performed with an in-house computer software named “Damızlık Asistanı” developed by Mustafa Tekerli̇. Correlations were calculated by Minitab 18.
The expected genetic gains (Venge and Christensen, 1969; Syrstad, 1970; Falconer and Mackay, 1996; Boareki et al., 2020) were predicted by the following equations:
where ΔGX is the direct response to selection on trait X; i is the intensity of selection, assumed to be equal to 1; is the direct additive heritability of trait X; and σPX is the phenotypic standard deviation of trait X.
Relative correlated responses in the ith trait were calculated with the following formula (Rao and Sundaresan, 1979):
Here, CRi is the correlated response of ith trait; rG(i,j) is the genetic correlation between traits i and j; hj is the square root of the heritability of jth trait against which selection is directed; and hi is the square root of the heritability of ith trait.
The effects of SNPs were tested with one-way ANOVA (Minitab 18) using the data adjusted for significant environmental effects.
The least-square means and ANOVA results for body measurements and ultrasound carcass traits of yearling Anatolian buffaloes are presented in Tables 1–2.
The farm had a significant (P<0.001) effect on all traits. Yılmaz et al. (2017), Erdoğan et al. (2021), Kaplan (2021), and Alkoyak and Öz (2022) reported similar results for the effects of village and rearing site on Anatolian buffaloes. The birth season was found to be significant (P<0.001) in only CW and HW. Sex had a significant (P<0.05) effect on BW, WH, BL, CBC, and LMD. Similar findings were obtained in different studies (Thiruvenkadan et al., 2009; Kul et al., 2017; Çelikeloğlu et al., 2019; Erdoğan et al., 2021; Kaplan, 2021; Alkoyak and Öz, 2022). BW values (175.41 ± 2.06 kg) were found to be higher than the results of earlier research (Thiruvenkadan et al., 2009; Akhtar et al., 2012; Şekerden, 2014; Pandya et al., 2015; Kul et al., 2017; Yılmaz et al., 2017; Erdoğan et al., 2021; Kaplan, 2021) for Murrah, Nili-Ravi, Surti, and Anatolian breeds, whereas higher values were reported by some studies (Shahin et al., 2010; Mendes Malhado et al., 2012; Vergara et al., 2012; Falleiro et al., 2013; Agudelo-Gómez et al., 2015; Shahjahan et al., 2017; Çelikeloğlu et al., 2019; El-den et al., 2020; Medrado et al., 2021). The least-square means for body measurements were similar to the results of literature (Uslu, 1965; Gürcan et al., 2011; Gavit et al., 2013; Çelikeloğlu et al., 2019). No information was encountered in the literature for ultrasound carcass traits of yearling buffaloes. This study was the first investigation in this context. Differences in the ANOVA may be due to age, breed, care, breeding, geography, and statistical models applied.
The variance components and direct additive, maternal, and total heritabilities estimated for the studied traits are shown in Table 3. The direct additive heritability for BW was found to be consonant with Medrado et al. (2021) and lower than the findings of Salces et al. (2006), Mendes Malhado et al. (2007), Shahin et al. (2010), Akhtar et al. (2012), Mendes Malhado et al. (2012), Vergara et al. (2012), Falleiro et al. (2013), Salces et al. (2013), Gupta et al. (2015), Pandya et al. (2015), El-den et al. (2020), and Kaplan (2021).
The estimate of direct additive heritability for WH was higher than that of Vankov and Peeva (1994). The direct heritability of rump height was higher than the findings in Nili-Ravi buffaloes (Mirza et al., 2020). The direct additive heritability estimates for BL were greater than those reported by Vankov and Peeva (1994) for yearlings and Thevamanoharan et al. (2001), and Mirza et al. (2020) for lactating buffaloes. The additive heritability for CC was 0.435 ± 0.040. While Vankov and Peeva (1994) reported a lower estimate for yearlings in this trait, Thevamanoharan et al. (2001) found a higher value in lactating buffaloes.
The estimate of heritability for LMA was lower than the result of Taborda et al. (2015) in buffaloes at 18 months of age. The data may be insufficient to detect a pronounced heritability estimate for LMA in our study. However, the direct additive heritability of SFT (0.539 ± 0.046) was larger according to the report of Taborda et al. (2015). The data frame and the statistical model may have caused differences between studies.
Maternal heritability for BW was found to be 0.646 ± 0.291 and higher than the results revealed by Vergara et al. (2012) and Falleiro et al. (2013) for Colombian and Mediterranean breeds. Buffalo breeders traditionally allow the calves to suckle only one teat of mothers during milking. Some high and significant maternal heritabilities obtained for body measurements and ultrasound carcass traits in our study may be an indicator the genetic capacity of mothers for milk production. Thus, Kushwaha et al. (2008) also emphasized the importance of maternal genetic effects, presumably reflecting differences in milk production.
The genetic and phenotypic correlations among the traits are presented in Table 4. All genetic and phenotypic correlations were significant (P<0.01) and ranged from moderate to high (except for CW and HW). These are the first results in connection with the body measurements and ultrasound carcass traits in the Anatolian buffalo population. No study was found discussing the genetic correlations among body size and ultrasound carcass traits in buffaloes.
The phenotypic correlations between BW and the other measurement traits were moderate to high (0.56–0.85) in the study. Opposite to these results, several authors (Sindhu and Pal, 1980; Paul and Das, 2012; Ahmad et al., 2013; Dhillod et al., 2017) reported positive but low to moderate correlations in different breeds. The correlations except for between CW and HW for body measurements were in the range of the literature (Paul and Das, 2012; Ahmad et al., 2013; Dhillod et al., 2017; de Melo et al., 2018; Nicolas et al., 2018; Dahiya et al., 2020; Ağyar et al., 2022). The significant and moderate genetic and phenotypic correlations between body measurements and ultrasound carcass traits indicated that newly integrated ultrasound techniques could improve the allometric traits. The correlations among ultrasound carcass traits themselves ranged from 0.53 to 0.83. These findings are in tune with the results of Andrighetto et al. (2010) for SFT and LMA. Accordingly, breeders may choose to measure muscle depth because of its practicality.
The direct and correlated responses between studied traits are presented in Table 5. The results showed that the most effective auxiliary trait is CC for improving others.
After sequencing analysis, SNPs investigated for PLAG1 and NCAPG genes were found to be monomorphic. In the LCORL gene, two genotypes were obtained. TT and TC genotypes were found in 227 and 9 animals with the means (BW) of 161.12 and 165.89 kg, respectively. CC (n: 162; : 161.12 kg), TC (n: 70; : 161.72 kg), and TT (n: 3; : 166.30 kg) genotypes were detected in the HMGA2 gene. The differences between genotypes in both genes were not significant.
This study is the first paper on genetic parameters in Anatolian buffalo for body size and ultrasound carcass traits. The genetic parameters obtained for body size and ultrasound carcass traits have shown that the Anatolian buffaloes could be genetically improved. The highest heritability was calculated in SFT, followed by WH, RH, BL, and CC. Genetic and phenotypic correlations among the traits were found to be in a desirable way and generally moderate to high. CC could be used as a criterion for indirect selection in animal improvement programs. Although there were no significant effects of SNPs in LCORL and HMGA2 genes, calves carrying genotypes of TC in LCORL and TT in HMGA2 had a tendency to be slightly heavier.
Data will be made available upon reasonable request.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by SÇ and MT. The first draft of the manuscript was written by SÇ and MT. All authors read and approved the final paper.
The contact author has declared that neither of the authors has any competing interests.
The animals were used in compliance with the rules of experimental animal ethics committee of the University of Afyon Kocatepe (decision nos. 49533702/258 and 210/20).
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
This study was summarized from the Ph.D. thesis of the first author. The authors are thankful to the Scientific Research Projects Committee of Afyon Kocatepe University and the General Directorate of Agricultural Research and Policies, Ministry of Agriculture and Forestry, Republic of Türkiye.
This research has been supported by Afyon Kocatepe Üniversitesi (grant nos. 49533702/258 and 210/20).
This paper was edited by Antke-Elsabe Freifrau von Tiele-Winckler and reviewed by Ö. Korkmaz Ağaoğlu and Banu Yüceer Ozkul.
Agudelo-Gómez, D., Pineda-Sierra, S., and Cerón-Muñoz, M. F.: Genetic Evaluation of Dual Purpose Buffaloes (Bubalus bubalis) in Colombia Using Principal Component Analysis, PLoS ONE, 10, e0132811, https://doi.org/10.1371/journal.pone.0132811, 2015.
Ağyar, O., Tırınk, C., Önder, H., Şen, U., Piwczyński, D., and Yavuz, E.: Use of Multivariate Adaptive Regression Splines Algorithm to Predict Body Weight from Body Measurements of Anatolian buffaloes in Türkiye, Animals, 12, 2923, https://doi.org/10.3390/ani12212923, 2022.
Ahmad, N., Abdullah, M., Javed, K., Khalid, M. S., Babar, M. E., Younas, U., and Nasrullah: Relationship between Body Measurements and Milk Production in Nili-Ravi Buffaloes Maintained at Commercial Farms in Peri-Urban Vicinity of Lahore, Buffalo Bull., 32, 792–795, 2013.
Akhtar, P., Kalsoom, U., Ali, S., Yaqoob, M., Javed, K., Babar, M. E., Mustafa, M. I., and Sultan, J. I.: Genetic and phenotypic parameters for growth traits of Nili-Ravi buffalo heifers in Pakistan, J. Anim. Plant Sci., 22, 347–352, 2012.
Alkoyak, K. and Öz, S.: The effect of nongenetic factors on calf birth weight and growth performance in Anatolian buffaloes, Turk. J. Vet. Anim. Sci., 46, 609–616, https://doi.org/10.55730/1300-0128.4232, 2022.
Andrighetto, C., Jorge, A. M., Francisco, C. D. L., Pinheiro, R. S. B., Silva, T. L., Surge, C. A., Andrade, C. R. M., Tavares, S. A., and Puoli Filho, J. N. P.: Relation between Ultrasound and Carcass Measures in Buffaloes Slaughtered in Different Stages of Feedlot, in: Proceedings of the 9th World Buffalo Congress, 25–28 April 2010, Buenos Aires, Argentina, 2010.
Arpacık, R.: Sığır yetiştiriciliği, Veteriner Fakültesi Yayınları, University of Uludağ, 1982.
Batu, S.: Umumi Zootekni, second edition, University of Ankara, Ankara, 1951.
Boareki, M. N., Brito, L. F., Cánovas, Á., Osborne, V., and Schenkel, F. S.: Estimation of genetic parameters and selection response for reproductive and growth traits in Rideau-Arcott sheep, Can. J. Anim. Sci., 101, 134–142, https://doi.org/10.1139/cjas-2019-0152, 2020.
Boom, R., Sol, C. J. A., Salimans, M. M. M., Jansen, C. L., Wertheim-Van Dillen, P. M. E., and Van Der Noordaa, J.: Rapid and simple method for purification of nucleic acids, J. Clin. Microbiol., 28, 495–503, https://doi.org/10.1128/jcm.28.3.495-503.1990, 1990.
Calo, L. L., McDowell, R. E., VanVleck, L. D., and Miller, P. D.: Genetic aspects of beef production among Holstein-Friesians pedigree selected for milk production, J. Anim. Sci., 37, 676–682, https://doi.org/10.2527/jas1973.373676x, 1973.
Çelikeloğlu, K., Koçak, S., Erdoğan, M., Bozkurt, Z., and Tekerli, M.: The investigation of viability and body measurements for water buffalo calves, Turk. J. Vet. Anim. Sci., 43, 60–67, https://doi.org/10.3906/vet-1809-44, 2019.
Çinkaya, S., Tekerli, M., Demirtaş, M., and Çelikeloğlu, K.: EasyNRM: A Visual Basic Application Approach for Computing Numerator Relationship Matrix of Pedigreed Animals, Kahramanmaraş Sütçü İmam Üniversitesi Tarım ve Doğa Dergisi, 22, 418–423, https://doi.org/10.18016/ksutarimdoga.vi.571575, 2019.
Dahiya, S. P., Kumar, M., and Dhillod, S.: Relationship of linear type traits with production and reproduction performancein Murrah buffaloes, Indian J. Anim. Sci., 90, 942–946, 2020.
de Melo, B. A., Nascimento, I. D. M., Santos, L. T. A. D., de Lima, L. G., de Araújo, F. C. T., Rios, R. R. S., Cuoto, A. D. G., and Fraga, A. B.: Body morphometric measurements in Murrah crossbred buffaloes (Bubalus bubalis), J. Appl. Anim. Res., 46, 1307–1312, https://doi.org/10.1080/09712119.2018.1502669, 2018.
Dhillod, S., Kar, D., Patil, C. S., Sahu, S., and Singh, N.: Study of the dairy characters of lactating Murrah buffaloes on the basis of body parts measurements, Veterinary World, 10, 17–21, https://doi.org/10.14202/vetworld.2017.17-21, 2017.
El-den, K., Mohammed, K. M., and Saudi, E. M.: Genetic and non-genetic factors affecting growth traits in Egyptian buffalo, Arch. Agr. Sci. J., 3, 1–12, https://doi.org/10.21608/AASJ.2020.41749.1032, 2020.
Erdoğan, M., Tekerli, M., Çelikeloğlu, K., Hacan, Ö., Kocak, S., Bozkurt, Z., Çinkaya, S., and Demirtaş, M.: Associations of SNPs in GHR gene with growth and milk yield of Anatolian buffaloes, Turkish J. Vet. Anim. Sci., 45, 1080–1086, https://doi.org/10.3906/vet-2103-40, 2021.
Falconer, D. S. and Mackay, T. F. C.: Introduction to quantitative genetics, Longman, Malaysia, ISBN: 0582243025, 1996.
Falleiro, V., Silveira, E. S., Ramos, A. A., Carneiro, P. L. S., Carrillo, J. A., and Malhado, C. H. M.: Genetic Parameters for Growth Traits of Mediterranean Buffaloes from Brazil, Estimated By Bayesian Inference, Buffalo Bull., 32, 627–631, 2013.
Gavit, D. P., Barbind, R. P., Korake, R. L., and Mule, P. R.: Studies on linear body measurement of Marathwadi buffalo on field scale, Res. J. Anim. Husband. Dair. Sci., 4, 10–12, 2013.
Gilbert, R. P., Bailey, D. R., and Shannon, N. H.: Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets, J. Anim. Sci., 71, 1712–1720, https://doi.org/10.2527/1993.7171712x, 1993.
Gudbjartsson, D. F., Walters, G. B., Thorleifsson, G., Stefansson, H., Halldorsson, B. V., Zusmanovich, P., Sulem, P., Thorlacius, S., Gylfason, A., Steinberg, S., Helgadottir, A., Ingason, A., Steinthorsdottir, V., Olafsdottir, E. J., Olafsdottir, G. H., Jonsson, T., Borch-Johnsen, K., Hansen, T., Andersen, G., Jorgensen, T., Pedersen, O., Aben, K. K., Witjes, J. A., Swinkels, D. W., den Heijer, M., Franke, B., Verbeek, A. L. M., Becker, D. M., Yanek, L. R., Becker, L. C., Tryggvadottir, L., Rafnar, T., Gulcher, J., Kiemeney, L. A., Kong, A., Thorsteinsdottir, U., and Stefansson, K.: Many sequence variants affecting diversity of adult human height, Nat. Genet., 40, 609–615, https://doi.org/10.1038/ng.122, 2008.
Gupta, J. P., Sachdeva, G. K., Gandhi, R. S., and Chakaravarty, A. K.: Developing multiple-trait prediction models using growth and production traits in Murrah buffalo, Buffalo Bull., 34, 347–355, 2015.
Gürcan, Y. T., Soysal, E. K., and Tuna, M. İ.: Anadolu mandalarının çeşitli vücut ölçülerine göre morfometrik karekterizasyonu, Tekirdağ Ziraat Fakültesi Dergisi, 8, 143–152, 2011.
Gürtan, K.: İstatistik ve Araştırma Metodları, Series of University of İstanbul, İstanbul, 1979.
Harada, H., Duran, P. G., Mamuad, F. V., Cruz, L. C., Domingo, I. J., and Ohashi, T.: Ultrasonic scanning for estimating the carcass traits of live water buffaloes, in: Proceedings of the 34. Annual Convention of the Philippine Society of Animal Science, 23–24 October 1997, Metro Manila, Filipinler, 1997.
Hwang, J. M., Cheong, J. K., Kim, S. S., Jung, B. H., Koh, M. J., Kim, H. C., and Choy, Y. H.: Genetic Analysis of Ultrasound and Carcass Measurement Traits in a Regional Hanwoo Steer Population, Asian-Austral. J. Anim., 27, 457–463, https://doi.org/10.5713/ajas.2013.13543, 2014.
İzmen, E. R. and Spöttel, W.: Anadolu Mandalarının Süt Verimleriyle Sütlerinin Terkibi, Yüksek Ziraat Enstitüsü Neşriyatı, 103, 1937.
Jorge, M., Andrighetto, C., Francisco, D. L., Neto, P., Mourão, D. C., Bianchini, W., Rodrigues, E., and Ramos, D. A.: Predicting beef carcass retail products of Mediterranean buffaloes by real-time ultrasound measures, Ital. J. Anim. Sci., 6, 1157–1159, https://doi.org/10.4081/ijas.2007.s2.1157, 2017.
Kaplan, Y.: Yozgat İli Anadolu Mandalarında Bazı Büyüme, Üreme ve Üretim Özelliklerini Etkileyen Genetik ve Çevresel Faktörlerin Tahmini, Ph.D. thesis, University of Afyon Kocatepe, Institute of Health Sciences, 115 pp., Afyonkarahisar, 2021.
Karim, L., Takeda, H., Lin, L. I., Druet, T., Arias, J. A., Baurain, D., Cambisano, N., Davis, S. R., Farnir, F., Grisart, B., Harris, B. L., Keehan, M. D., Littlejohn, M. D., Spelman, R. J., Georges, M., and Coppieters, W.: Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature, Nat. Genet., 43, 405–413, https://doi.org/10.1038/ng.814, 2011.
Kendir, H. S.: Çifteler harasıve Eskişehir bölgesi halk elindeki bozkır X montafon melezi sığırların form beden ölçüleri ve başlıca verimleri üzerinde araştırma, Ph.D. thesis, University of Ankara, 176 pp., Ankara, 1960.
Koçak, S., Tekerli, M., Çelikeloğlu, K., Erdoğan, M., and Hacan, Ö.: An investigation on yield and composition of milk, calving interval and repeatabilities in riverine buffaloes of Anatolia, J. Anim. Plant Sci., 29, 650–656, 2019.
Korejo, N. A., Parveen, F., Memon, M. I., Legharı, R. A., Sethar, A., Kumbhar, H. K., Memon, M. R., Kumbhar, S. N., Korejo, R. A., Channa, A. A., and Kalwar, Q.: Age-related changes in body weight, Body Conformation and Scrotal Circumference and Prepubertal Sexual behavior of Kundhi Buffalo Bull Calves, Sindh University Research Journal-SURJ, 51, 75–80, 2019.
Kul, E., Şahin, A., Çayıroğlu, H., Filik, G., Uğurlutepe, E., and Kaplan, Y.: Non Genetic Factors Affecting Some Growth Traits in Anatolian Buffaloes, in: Proceedings of the 8th Balkan Animal Science Conference BALNIMALCON 2017, 6–8 September 2017, Prizren, Kosovo, 2017.
Kushwaha, B. P., Mandal, A., Arora, A. L., Kumar, R., Kumar, S., and Notter, D. R.: Direct and maternal (co) variance components and heritability estimates for body weights in Chokla sheep, J. Anim. Breed. Genet., 126, 278–287, 2009.
Mendes Malhado, C. H., Ramos, A. A., Carneiro, P. L. S., de Souza, J. C., and Lamberson, W. R.: Genetic and phenotypic trends for growth traits of buffaloes in Brazil, Ital. J. Anim. Sci., 6, 325–327, https://doi.org/10.4081/ijas.2007.s2.325, 2007.
Mendes Malhado, C. H., Mendes Malhado, A. C., Amorim Ramos, A., Souza Carneiro, P. L., Siewerdt, F., and Pala, A.: Genetic parameters by Bayesian inference for dual purpose Jaffarabadi buffaloes, Arch. Anim. Breed., 55, 567–576, https://doi.org/10.5194/aab-55-567-2012, 2012.
Medrado, B. D., Pedrosa, V. B., and Pinto, L. F. B.: Meta-analysis of genetic parameters for economic traits in buffaloes, Livest. Sci., 251, 104614, https://doi.org/10.1016/j.livsci.2021.104614, 2021.
Meyer, K.: Variance components due to direct and maternal effects for growth traits of Australian beef cattle, Livest. Prod. Sci., 31, 179–204, https://doi.org/10.1016/0301-6226(92)90017-X, 1992.
Meyer, K.: WOMBAT – A tool for mixed model analyses in quantitative genetics by restricted maximum likelihood (REML), J. Zhejiang Univ.-Sc. B, 8, 815–821, https://doi.org/10.1631/jzus.2007.B0815, 2007.
Mirza, R. H., Khan, M. A., Waheed, A., Akhtar, M., Raihan-Dilshad, S. M., Asim, F., Ishaq, H. M., and Kuthu, Z. H.: Heritability estimates of some body measurements in Nili Ravi buffaloes, J. Anim. Plant Sci., 30, 537–544, https://doi.org/10.36899/JAPS.2020.3.0063, 2020.
Mitchell, R. G., Rogers, G. W., Dechow, C. D., Vallimont, J. E., Cooper, J. B., Sander-Nielsen, U., and Clay, J. S.: Milk urea nitrogen concentration: heritability and genetic correlations with reproductive performance and disease, J. Dairy Sci., 88, 4434–4440, https://doi.org/10.3168/jds.S0022-0302(05)73130-1, 2005.
Nicolas, F. F. C., Saludes, R. B., Relativo, P. L. P., and Saludes, T. A.: Estimating live weight of Philippine dairy buffaloes (Bubalus bubalis) using digital image analysis, Philipp. J. Vet. Anim. Sci., 44, 129–138, 2018.
Pandya, G. M., Joshi, C. G., Rank, D. N., Kharadi, V. B., Bramkshtri, B. P., Vataliya, P. H., Desai, P. H., and Solanki, J. V.: Genetic analysis of body weight traits of Surti buffalo, Buffalo Bull., 34, 189–195, 2015.
Paul, S. S. and Das, K. S.: Prediction of body weight from linear body measurements in Nili-Ravi buffalo calves, J. Buff. Sci., 1, 32–34, https://doi.org/10.6000/1927-520X.2012.01.01.06, 2012.
Peeva, T.: Estimation of type and body conformation in buffaloes, in: Proceedings of the 3rd World Buffalo Congress, 13–17 May 1991, Varna, Bulgaria, 1991.
Pryce, J. E., Hayes, B. J., Bolormaa, S., and Goddard, M. E.: Polymorphic regions affecting human height also control stature in cattle, Genetics, 187, 981–984, https://doi.org/10.1534/genetics.110.123943, 2011.
Rao, M. K. and Sundaresan, D.: Influence of environment and heredity on the shape of lactation curves in Sahiwal cows, J. Agr. Sci., 92, 393–401, https://doi.org/10.1017/S0021859600062924, 1979.
Rebak, G., Capellari, A., Rivolta, A. Y., and Alarcón, A.: Exploratory study of ultrasound on properties of meat in buffaloes in the northeast of Argentina, in: Proceedings of the 9th World Buffalo Congress, 25–28 April 2010, Buenos Aires, Argentina, 2010.
Riaz, R., Tahir, M. N., Waseem, M., Asif, M., and Khan, M. A.: Accuracy of estimates for live body weight using Schaeffer's formula in non-descript cattle (bos indicus), nili ravi buffaloes (bubalus bubalis) and their calves using linear body measurements, Pakistan J. Sci., 70, 225–232, 2018.
Şahin, A., Ulutaş, Z., and Yıldırı m, A.: Türkiye ve Dünya'da Manda Yetiştiriciliği, Gaziosmanpaşa Bilimsel Araştırma Dergisi, 8, 65–70, 2013.
Salces, C. B., Salces, A. J., Seo, K. S., and Bajenting, G. P.: Genetic parameter estimation of growth traits of Murrah Buffaloes raised under ranch production system, in: Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, 13–18 August 2006, Belo Horizonte, Brazil, 2006.
Salces, A. J., Bajenting, G. P., and Salces, C. B.: Estimation of genetic parameters for growth traits of three genotypes of water buffalo bulls raised on a ranching operation, Buffalo Bull., 32, 760–763, 2013.
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W.: NIH Image to ImageJ: 25 years of image analysis, Nat. Method., 9, 671–675, https://doi.org/10.1038/nmeth.2089, 2012.
Şekerden, Ö.: Growth traits of Anatolian and Anatolian × Italian crossbred buffalo calves under the village conditions, J. Buff. Sci., 3, 92–96, https://doi.org/10.6000/1927-520X.2014.03.03.5, 2014.
Setoguchi, K., Furuta, M., Hirano, T., Nagao, T., Watanabe, T., Sugimoto, Y., and Takasuga, A.: Cross-breed comparisons identified a critical 591-kb region for bovine carcass weight QTL (CW-2) on chromosome 6 and the Ile-442-Met substitution in NCAPG as a positional candidate, BMC Genet., 10, 1–12, https://doi.org/10.1186/1471-2156-10-43, 2009.
Shahin, K. A., Abdallah, O. Y., Fooda, T. A., and Mourad, K. A.: Selection indexes for genetic improvement of yearling weight in Egyptian buffaloes, Arch. Anim. Breed., 53, 436–446, https://doi.org/10.5194/aab-53-436-2010, 2010.
Shahjahan, M., Khatun, A., Khatun, S., Hoque, M. M., Hossain, S., Huque, Q. M. E., Awal, T. M., and Mintoo, A. A.: Study on growth traits at weaning and yearling stages of indigenous and F1 crossbred buffalo in Bangladesh, Asian J. Med. Biol. Res., 3, 499–503, https://doi.org/10.3329/ajmbr.v3i4.35341, 2017.
Sindhu, M. S. and Pal, R. N.: Phenotypic correlations of external body measurements with body weight, lactation yield and fat and solid content of milk in Murrah buffaloes [India], Haryana Agricultural University Journal of Research, 10, 112–114, 1980.
Singh, P. K., Ahlawat, S. S., Sharma, D. P., and Pathera, A.: Carcass characteristics of male buffalo calf and meat quality of its veal, Buffalo Bull., 37, 129–144, 2018.
Soysal, M. İ.: Manda ve Ürünleri Üretimi, Unpublished lecture notes, University of Namık Kemal, Faculty of Agriculture, 2009.
Syrstad, O.: Estimating Direct and Correlated Response to Selection. A Note for Clarification, Acta Agr. Scand., 20, 205–206, https://doi.org/10.1080/00015127009433408, 1970.
Taborda, J. J., Cerón-Muñoz, M. F., Barrera, D. C., Corrales, J. D., and Agudelo, D. A.: Inferencia bayesiana de parámetros genéticos para características de crecimiento en búfalos doble propósito en Colombia, Livestock Research for Rural Development, 27, 196, http://www.lrrd.org/lrrd27/10/cero27196.html (last access: 14 November 2023), 2015.
Thevamanoharan, K., Vandepitte, W., Mohiuddin, G., Chantalakhana, C., and Bunyavejchewin, P.: Genetic, phenotypic and residual correlations among various growth traits and between growth traits and body measurements of Swamp buffaloes, Buffalo Bull., 19, 55–60, 2000.
Thiruvenkadan, A. K., Panneerselvam, S., and Rajendran, R.: Non-genetic and genetic factors influencing growth performance in Murrah Buffalos, S. Afr. J. Anim. Sci., 39, 102–106, https://doi.org/10.4314/sajas.v39i1.61326, 2009.
Uslu, N. T.: 1963–1965 yıllarında Afyon Yem Bitkileri Üretme ve Zootekni Deneme İstasyon Müdürlüğünde ve Çevresindeki Mandacılık, Tarım Bakanlığı Ziraat İşleri Genel Müdürlüğü Yayınları, Yayın no: D – 114, Afyon, 1965.
Uslu, N. T.: Afyon Bölgesi Mandalarının Çeşitli Özellikleri ile Rasyonel ve Köy Şartlarında Süt Verimleri Üzerine Mukayeseli Araştırmalar, Ph.D. thesis, Faculty of Agriculture, University of Egean, Bornova, İzmir, 81 pp., 1970.
Vankov, K. and Peeva, T.: Genetic parameters of body measurements in buffalo calves, in: Proceedings of the 4th World Buffalo Congress, 27–30 June 1994, Sao Paulo, Brazil, 1994.
Venge, O. and Christensen, K.: Expected Genetic Gain in Amount of Protein by Selection for Butterfat in Red Danish Cattle, Acta Agr. Scand., 19, 99–102, https://doi.org/10.1080/00015126909433397, 1969.
Vergara, D. M. B., Muñoz, M. F. C., Toro, E. J. R., Gómez, D. A. A., and Cifuentes, T.: Genetic parameters for growth traits of buffaloes (Bubalus bubalis Artiodactyla, Bovidae) in Colombia, Rev. Colomb. Cienc. Pec., 25, 202–209, 2012.
Yılmaz, A., Ocak, E., and Köse, S.: A research on milk yield, milk composition and body weights of Anatolian buffaloes, Indian J. Anim. Res., 51, 564–569, https://doi.org/10.18805/ijar.11474, 2017.





