the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Single-nucleotide polymorphisms in FLT3, NLRP5, and TGIF1 are associated with litter size in Small-tailed Han sheep
Si Chen
Lin Tao
Xiaoyun He
Ran Di
Xiangyu Wang
Mingxing Chu
Previous studies have indicated that FLT3, NLRP5, and TGIF1 play a pivotal role in sheep fecundity. Nevertheless, little is known about the association of the polymorphisms of these genes with litter size (LS). In this study, the selected single-nucleotide polymorphisms (SNPs) were genotyped using a Sequenom MassARRAY® platform, and the distribution of different genotypes of the SNPs in the seven sheep breeds (Small-tailed Han, Hu, Cele Black, Suffolk, Tan, Prairie Tibetan, and Sunite sheep) were analyzed. The reliability of the estimated allele frequency for all seven SNPs was at least 0.9545. Given the association of the TGIF1 g.37866222C > T polymorphism with LS in Small-tailed Han sheep (p<0.05), fecundity differences might be caused by the change in amino acid from proline (Pro) to serine (Ser), which has an impact on secondary, tertiary protein structures with concomitant TGIF1 functionality changes. The FLT3 rs421947730 locus has a great effect on the LS (p<0.05), indicating that the locus of FLT3 in synergy with KILTG is likely to facilitate ovarian follicle maturation and ovulation. Moreover, NLRP5 rs426897754 is associated with the LS of the second and third parities (p<0.05). We speculate that a synonymous variant of NLRP5 may be involved in folliculogenesis accompanied by BMP15, FSHR, BMPR1B, AMH, and GDF9, resulting in the different fecundity of Small-tailed Han sheep. Our studies provide valuable genetic markers for sheep breeding.
- Article
(4212 KB) - Full-text XML
- BibTeX
- EndNote
Many physiological parameters involved in animal reproduction have an important role in the productivity of farm animals. Advances in research on reproduction have enabled the identification of strategies to improve reproductive and productive capacity in livestock. For instance, it has been established that melatonin administration can be used to modify seasonal breeding patterns in goats and sheep (Deveson et al., 1992, Mura et al., 2010) or to decrease milk yield by altering prolactin concentrations in lactating ewes (Molik et al., 2013). Moreover, available evidence suggests that as day length decreases during the autumn, an increase in the duration of melatonin secretion triggers an endocrine response, leading to the onset of reproductive activity (Chemineau et al., 1987). Manipulation of photoperiod and melatonin treatment are widely used for the control of reproduction in goats and sheep (Carcangiu et al., 2015, Giannetto et al., 2020, 2021). Fecundity, one of the significant economic indicators on sheep breeds, has a direct effect on the economic benefit of sheep. The measure of sheep fertility is the litter size (LS) of ewes; thus, increasing the LS plays a pivotal role in developing the sheep industry. The lambing character is quantitative in nature and is controlled by minor polygenes. Molecular markers associated with a quantitative trait can select the genotype directly. That is, combining traditional selection with modern molecular breeding technology greatly boosts the efficiency of sheep breed selection, further improving reproductive performance (Notter, 2012).
During the last decade, microsatellites have been the marker of choice for most parentage and assignment studies because of their high variability and wide availability. Microsatellites are DNA regions composed of a short sequence of two to six nucleotides repeated several times that show high levels of intraspecific allele polymorphism and are widely distributed within genomes of both terrestrial and marine vertebrates (Dekkers, 2014, Arfuso et al., 2017). Furthermore, single-nucleotide polymorphisms (SNPs), which identify the polymorphisms of single nucleotides, are considered useful molecular markers in several research fields, such as LS (Mahdavi et al., 2014, Chong, et al., 2018, Wang et al., 2018).
FLT3 (Fms Related Receptor Tyrosine Kinase 3) mainly exists in haematopoietic stem cells to regulate hematopoiesis (Lyman and Jacobsen, 1998). Upon stimulation with the FLT3 ligand, the increasing activity of FLT3 triggers intracellular signaling, such as the PI3K cascade, promoting preimplantation embryo development (Nishijima et al., 2014). In contrast to wild-type FLT3 signaling, mutated FLT3 potently triggers the STAT5 pathway (Takahashi, 2011). STAT5 induces cycling D1, antiapoptotic gene p21, and c-myc to be involved in cell growth (Takahashi et al., 2004). These effects may indicate the roles of SNPs of FLT3 in the reproductive traits, such as LS. A recent study reported that the FLT3 gene is expressed in oocytes, and its protein is expressed on the oocyte membrane, indicating that FLT3 modulates follicle growth and maturation similar to CSF-1R and c-kit (Okamura et al., 2001).
NLRP5 (NLR Family Pyrin Domain Containing 5), as a maternal-effect gene, is essential for embryonic progress and female fertility (Velummailum, 2011). Female mice with NLRP5 deletion undergo normal process from oogenesis and folliculogenesis to ovulation (Tong et al., 2004). In addition, they normally respond to the stimulation of exogenous gonadotropin, producing a similar number and morphology of oocytes to wild type animals (Tong et al., 2004). Nevertheless, NLRP5-deficient mice have meiotic spindle defects in oocytes, which may increase DNA damage (Ohsugi et al., 2008). Therefore, NLRP5 may be an integral component that functions to preserve the genome integrity of female gametes (Velummailum, 2011).
TGIF1 (TGFβ Induced Factor Homeobox 1) has been reported to mediate the gonadotropin-releasing hormone (GnRH) pulse sensitivity of FSH-β (Mistry et al., 2011). Under a slow GnRH pulse frequency, overexpressed corepressor TGIF1 suppress the activation of the FSH-β promoter but has little effect on the fast pulse frequency (Hyman et al., 2003). On the contrary, knockout TGIF1 selectively increases the expression level of FSH-β mRNA on the fast GnRH pulse frequency (Mistry et al., 2011); that is, TGIF1 modulates FSH synthesis and secretion in different ways. Moreover, TGIF1 is highly expressed in the sheep ovary and functions as a transcriptional repressor on the SMAD-induced TGF-β signaling in vertebrates (Jiang et al., 2014, Guca et al., 2018), indicating that TGIF1 may be involved in reproductive process.
To reveal the relationship of the polymorphism of FLT3, NLRP5, and TGIF1 with LS, this study intends to screen out the loci of SNPs in the seven abovementioned native Chinese sheep breeds and genotype them using a Sequenom MassARRAY® platform. By analyzing the distribution of various genotypes on these SNPs in various sheep groups, it is expected to obtain the loci of the SNPs related to reproductive traits and provide valuable genetic markers for sheep genetics and breeding.
2.1 Animal, sample preparation, and data measurement
All experimental procedure involving sheep were supported by the Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China). Ethics approval was offered by the animal ethics committee of IAS-CAAS (no. IAS2020-63).
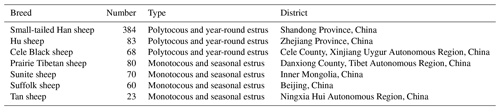
As shown in Table 1, a total of 768 sheep aged around 3 years were selected, including three higher-fecundity breeds (Small-tailed Han sheep with three consecutive lambing periods, n=384; Hu sheep, n=83; Cele Black sheep, n=68) and four lower-fecundity breeds (Suffolk sheep, n=60; Tan sheep, n=23; Prairie Tibetan sheep, n=80; Sunite sheep, n=70). Blood sampling was performed at the same time. Blood samples were collected into ethylenediaminetetraacetic-acid-coated tubes (Rongxin Bio-Tech Co., Ltd., Beijing, China) via jugular vein catheters (10 mL of blood from each animal) and were stored at −20 ∘C for further research. The lambing season, number of births, and litter size were then recorded in turn.
2.2 DNA extraction and detection
DNA extraction was carried out as per the blood genomic DNA extraction kit (Tiangen Biochemical Technology Co) instructions. The purity and concentration of DNA were detected using a NanoDrop 2000 spectrophotometer, and the tissue DNA's integrity was tested using 1.5 % agarose gel electrophoresis.
2.3 Design of the primer and synthesizing and genotyping of the locus
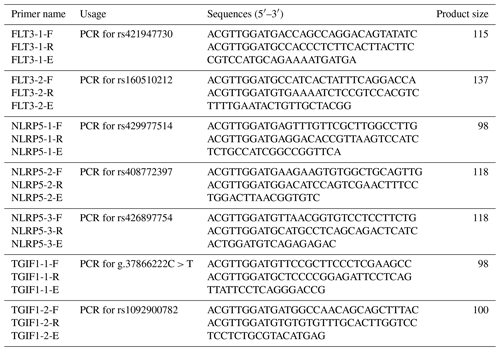
Combined with our previous data on 768 sheep from seven breeds, which was collected via whole-genome resequencing, some SNPs were screened out (Zhang et al., 2019, 2020; He et al., 2019), including rs421947730 and rs160510212 of FLT3; rs429977514, rs408772397, and rs426897754 of NLRP5; and g.37866222C > T and rs1092900782 of TGIF1. Primers to genotype these SNPs were designed by MassARRAY® Assay Design (v.3.1). MassARRAY® is a strategy for the detection of SNPs, including amplifying target sequences and performing single-base extended polymerase chain reactions (PCRs) with a specific primer (Ellis and Ong, 2017). Due to the fact that various bases occur at polymorphic sites, the amplified products have differences in their molecular weight following extension. These differences enable the products to be separated on the basis of genotype using time-of-flight mass spectrometry. The primers were synthesized by Beijing Tianyi Huiyuan Biotechnology Co. The primer sequences are shown in Table 2.
2.4 Statistical analysis
PopGene32 (version 3.2) software and the PIC-CALC procedure were used to process the results obtained from SNP genotyping and to calculate the PIC (polymorphism information content), HE (heterozygosity), and p values (χ2 test). The particular sheep population, with p>0.05 (χ2 test), was regarded to conform to a Hardy–Weinberg equilibrium (Hui and Burt, 2020). The following animal model was applied: . Here, yijkl is the phenotypic value observed in lambing; μ refers to the average population; Genotypei is the effect of the ith genotypes (); Pj represents the effect of the jth parity (); Ik indicates the parity–genotype interaction effect; and eijkl is the symbol of random errors, which are supposed to be independent of each other and follow a distribution.
The reliability was calculated using the formulas given in the following, ensuring that the estimation of allele frequencies did not deviate from the true value by more than 0.5 times and ensuring the relative deviation when the reliability was at least 0.9545 (Chang et al., 2000).
Here, β, η, λ, P, and V(P) stand for the reliability that allele frequencies do not deviate from the true value more than 0.5 times, the relative deviation when the reliability is at least 0.9545, the standard deviation from the estimate, gene frequencies, and gene variances, respectively.
2.5 Predictive protein interaction work of the target genes
Figuring out the protein–protein interactions contributes to deeply understanding the molecular mechanism of FLT3, NLRP5, and TGIF1 for complex reproductive traits, such as the LS. The respective transmembrane domains in FLT3, NLRP5, and TGIF1 were predicted by TMHMM Server (v.2.0, https://services.healthtech.dtu.dk/service.php?TMHMM-2.0, last access: March 2021). Prediction of the secondary structure, solvent accessibility, ConSurf (Ashkenazy, 2016), and binding site of TGIF1 were carried out using PredictProtein (https://www.predictprotein.org/, last access: March 2021). The STRING database (v.11.0) was applied to collect and integrate these functional interactions by consolidating protein–protein association data for massive organisms.
3.1 Blood DNA test results
The NanoDrop 2000 and 1.5 % agarose gel electrophoresis were used to detect the purity, concentration, and integrity of tissue DNA from the seven respective breeds. The test results showed ratios of D260 nmD280 nm between 1.8 and 2.0, all concentrations above 30 ng µL−1, and qualified electrophoresis bands; that is, the results could be used for further analysis.
3.2 Position mutation analysis of SNPs in the FLT3, NLRP5, and TGIF1 genes
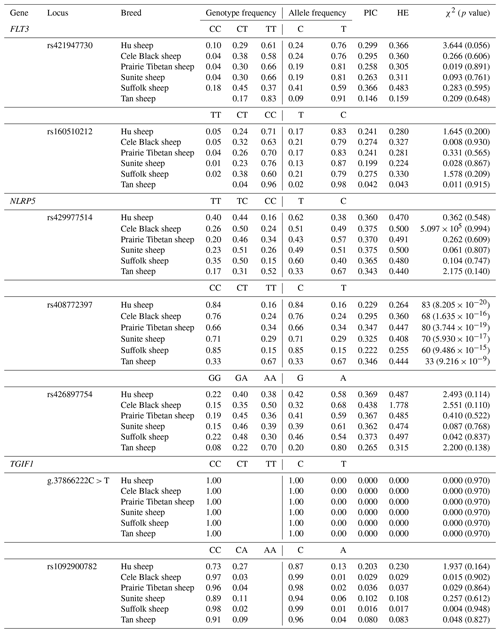
Seven potential SNPs were located in the exon region, including six synonymous variants and one missense variant. Among them, the number of A/G transitions, T/C transitions, and C/A transversions was 1, 5, and 1, respectively, and the phenomenon of T/A, G/C, and G/T transversion was not revealed. The number of transitions accounted for 85.71 % of the total detected SNPs, whereas transversions accounted for 14.29 % (Fig. 1). According to the genotyping results, the FLT3 gene rs421947730 and rs160510212 possessed two genotypes (CT, TT) in the Tan sheep with low fecundity but three genotypes (CC, CT, TT) in the other six sheep breeds. The NLRP5 gene rs408772397 possessed two genotypes (CC, TT) in the seven sheep breeds. The other two SNPs of NLRP5 had three genotypes in all experimental sheep breeds. The TGIF1 gene g.37866222C > T had two genotypes (CC, CT) in the Small-tailed Han sheep but one genotype (CC) in the other five breeds. The other SNP rs1092900782 had two genotypes (CC, CA) in the seven sheep breeds (Fig. 1, Table 3).
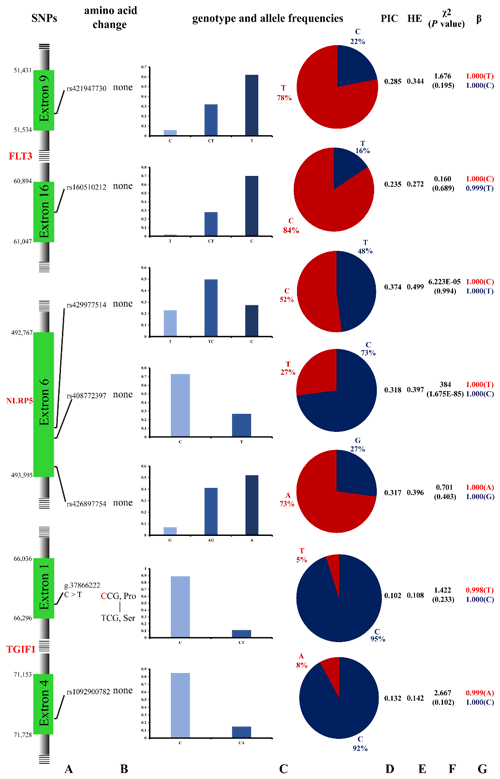
Figure 1Polymorphisms of the seven SNPs from the FLT3, NLRP5, and TGIF1 genes in Small-tailed Han sheep: (a) the position of seven SNPs on the chromosome; (b) the change in amino acids; (c) the genotype and allele frequencies of the FLT3, NLRP5, and TGIF1 genes; (d) the polymorphism information content (PIC) test; (e) the heterozygosity (HE) test; and (f) the Hardy–Weinberg equilibrium (HWE) test, , where O and E are observed frequency and expected frequency, respectively. (g) β represents the reliability of the estimated allele frequencies.
3.3 Population genetic analysis in the six sheep breeds
Besides Small-tailed Han sheep, the population genetic results of seven SNPs in the other six sheep breeds (Hu, Cele Black, Prairie Tibetan, Sunite, Suffolk, and Tan sheep) are shown in Table 3. In the FLT3 gene, the rs421947730 locus had low polymorphism (PIC≤0.25) in Tan sheep but moderate polymorphism () in the other sheep breeds; it was also under Hardy–Weinberg equilibrium (HWE, p>0.05) in all of the sheep breeds. The rs160510212 locus had moderate polymorphism in Cele Black and Suffolk sheep but low polymorphism in other groups; it also corresponded to the HWE in all groups. In the NLRP5 gene, both the rs429977514 and rs426897754 loci had moderate polymorphism and were under HWE in the six sheep breeds. The rs408772397 sites had low polymorphism in Hu and Suffolk sheep but moderate polymorphism in the residual sheep breeds; these loci significantly deviated from HWE in all of the sheep breeds (p≤0.05). In the TGIF1 gene, the g.37866222C > T and rs1092900782 loci had low polymorphism and were under HWE in the six sheep breeds (Table 3). Moreover, the reliability of estimated allele frequencies was at least 0.9545 in all cases.
3.4 Polymorphism analysis of the locus in the Small-tailed Han sheep
As is shown in the Fig. 1, the reliability of the estimated allele frequencies was at least 0.954 in all cases. The χ2 test indicated that one SNP (rs408772397) significantly deviated from Hardy–Weinberg equilibrium (p≤0.05). Moreover, the SNP g.37866222C > T, whose base C is replaced by base T, resulting in proline (Pro) substituted for serine (Ser), is described for the first time in this study.
3.5 The association of FLT3, NLRP5, and TGIF1 polymorphisms with litter size in Small-tailed Han sheep
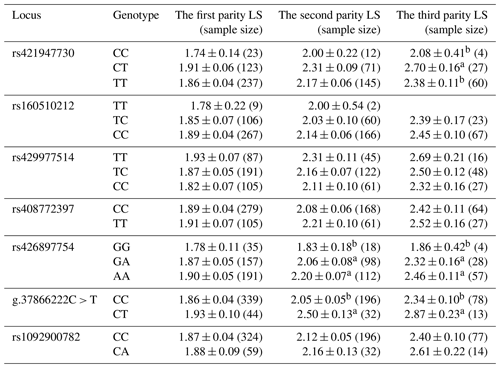
Litter sizes (LSs) with three parities of different loci were recorded in this study (Table 4). In the first parity, the LS with the mutant type was greater than that for the wild type at most loci. The rs421947730 locus of FLT3 was associated with the LS at the third parity, and ewes with the CT variant had the highest lambing rate (p<0.05). The rs426897754 locus of NLRP5 was related to the LS at the second and third parity, and mutant-type sheep with GA had the highest lambing rate (p<0.05). The g.37866222C > T locus of TGIF1 was associated with the LS at the second and third parity, and mutant-type ewes (CT) were associated with the largest LS (p<0.05); that is, mutant heterozygote ewes produced the maximum LS in our results.
3.6 Bioinformatic analysis of FLT3, NLRP5, and TGIF1
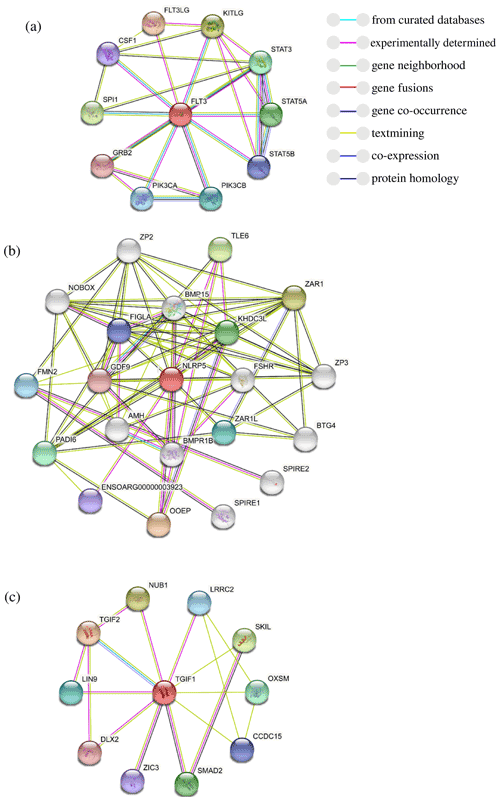
To further explore the changes before and after the mutation of genes, some bioinformatic methods were applied, such as transmembrane domain prediction (TMHMM), protein prediction (PredictProtein), and functional protein association networks (STRING). The amino acid residues 1–531 were located outside, 532–554 were located in the transmembrane region, 555–663 were located inside, 664–686 were located in the transmembrane region, and 687–983 were located outside of the FLT3 protein membrane. The synonymous variants of rs421947730 and rs160510212 were set outside of the FLT3 protein membrane and the transmembrane regions, respectively. Moreover, the amino acid residues 1–1200 and 1–406 were located outside of the NLRP5 and TGIF1 protein membrane, respectively (Fig. 2). That is, the replacement of Pro with Ser at position 63 of g.37866222C > T occurred outside of the TGIF1 protein membrane. The analysis of protein prediction indicated that the obvious differences in TGIF1 g.37866222C > T existed in the secondary structure, solvent accessibility, ConSurf, and binding site prediction (Fig. 3). This missense variant induced a change in the surrounding sites from average or conserved to variable and also increased the number of protein-binding sites, RNA-binding sites, and DNA-binding sites, indicating that g.37866222C > T might affect TGIF1 function with respect to reproduction. Protein interaction networks indicated that FLT3, NLRP5, and TGIF1 were predicted to interact with the known proteins affecting the ovulation rate and LS, such as FLT3 with KITLG; NLRP5 with BMP15, GDF9, AMH, BMPR1B, and FSHR; and TGIF1 with SMAD2 (Fig. 4).
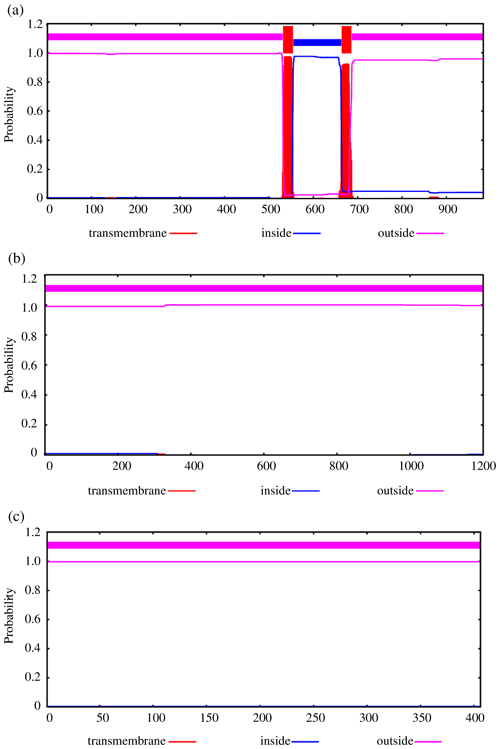
Figure 2Transmembrane regions' prediction using the amino acid sequence reported in TMHMM, including FLT3 (a), NLRP5 (b), and TGIF1 (c).
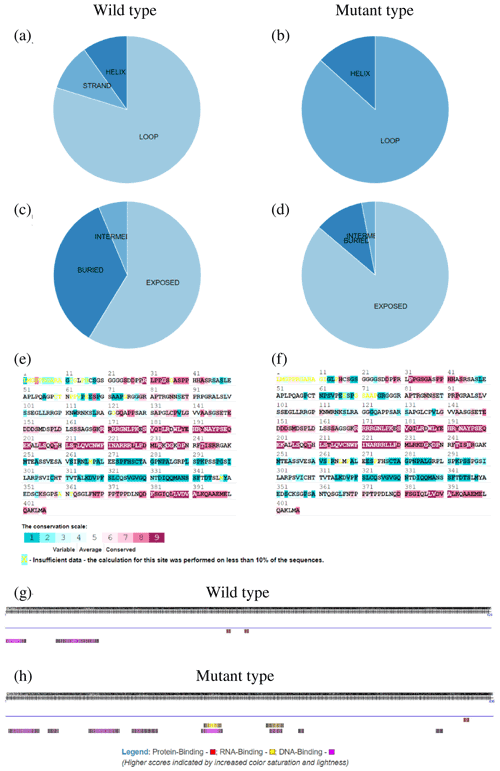
Figure 3Secondary protein structure for the TGIF1 product at g.37866222C > T based on its predicted amino acid sequence: (a, b) secondary structure composition of wild and mutant type, respectively; (c, d) solvent accessibility of wild and mutant type, respectively; (e, f) ConSurf results of wild and mutant type, respectively; and (g, h) binding site prediction of wild and mutant type, respectively.
Marker-assisted selection with traditional selection is potentially effective for the improvement of traits that are of low heritability, such as ovulation rate and LS (Williams, 2005). FLT3 and NLRP5 are both expressed in oocytes and are involved in follicle growth and maturation to increase the ovulation rate (Okamura et al., 2001, Bebbere et al., 2014, Velummailu, 2011). TGIF1, a SMAD transcriptional corepressor, negatively dictates the TGF-β signaling pathway to repress reproduction (Guca et al., 2018). The rs421947730 and rs160510212 loci of FLT3, rs408772397 of NLRP5, and g.37866222C > T and rs1092900782 of TGIF1 exhibited low polymorphism (PIC≤0.25), which may be limited by the sample size. Due to selection, several sheep breeds were not in HWE (p≤0.05) at these loci. This study reveals that some SNPs of these genes are associated with the LS in Small-tailed Han sheep.
Missense mutations have a great effect on complex traits. Point mutations in GDF9 (Tong, 2020), BMPR1B (Souza, 2001), and BMP15 (Hanrahan, 2004) have been reported to significantly affect the LS in sheep. Similar to previous studies, our study revealed that the TGIF1 missense variant g.37866222C > T showed a significant increase in LS at the second and third parity and that mutant-type ewes (CT) displayed the maximum LS (p<0.05). However, the mutant homozygous genotype TT was not detected, similar to g.37871539C > T in TGIF1 (Zhang, 2020), indicating that lethality may be caused by mutant homozygous genotypes at some loci of TGIF1 in all sheep breeds. Moreover, cellular transmembrane domains enable numerous proteins to function on the fertility-related signaling pathway (Guna and Hegde, 2018). In our results, the replacement of Pro with Ser at g.37866222C > T occurred outside of the TGIF1 protein membrane, indicating that g.37866222C > T might be a ligand-binding site. Given that TGIF1 interacts with SMAD2 via protein–protein networks, the missense variant decreases critical protein-binding sites, thereby weakening the formation of the protein complex. Once the protein complex has been reduced, the TGIF1–SMAD system would not be released from promoters or enhancers, resulting in TGIF1 being unable to function on TGF-β signaling as an active repressor of SMAD complexes (Guca et al., 2018). Considering the association of g.37866222C > T polymorphism with LS in Small-tailed Han sheep, fecundity differences might be caused by the change in amino acid from Pro to Ser, which has an impact on secondary and tertiary protein structures, causing TGIF1 functionality changes.
The FLT3 gene has been demonstrated to distribute on oocytes, and its protein distributes on the membrane of oocytes, indicating that FLT3 is involved in various stages of folliculogenesis like c-kit and CSF-1R (Okamura et al., 2001). In the protein–protein networks, FLT3 is predicted to interact with KITLG. In the ovine follicles, KITLG mRNA is highly expressed in early antral follicles but reduced at the late antral stage with follicular growth progress (Joyce et al., 2000). KITLG–KIT interactions exist in several stages of follicular development (Yoshida et al., 1997), and they function on ovarian follicle maturation (An et al., 2015). Thus, KITLG may be a candidate gene for the selection of sheep reproductive traits. The rs421947730 locus has a significant difference in the litter size of mutant type (CT, TT) versus wild type (CC), indicating that the locus of FLT3 in synergy with KITLG is likely to facilitate ovarian follicle maturation and ovulation in Small-tailed Han sheep. NLRP5 wild- and mutant-type females have similar follicular recruitment rates and a similar ovarian germ pool. Nevertheless, ovulated oocytes with NLRP5 deletion induce premature activation of the mitochondria, followed by embryo demise due to mitochondrial depletion (Fernandes et al., 2012). In the analysis of protein prediction, NLRP5 protein interacts with the proteins related to follicular development, such as BMP15, FSHR, BMPR1B, AMH, and GDF9 (Chu et al., 2007; Feary, et al., 2007). Moreover, at the rs426897754 locus of NLRP5, the LS of mutant-type (GA, AA) sheep is higher than that for wild-type (GG) sheep at the second and third parity. We suggest that NLRP5 with a synonymous variant may be involved in folliculogenesis accompanied by BMP15, FSHR, BMPR1B, AMH, and GDF9, resulting in the different fecundity of Small-tailed Han sheep.
Among these SNPs, g.37866222C > T of TGIF1 at the second and third parity, rs421947730 of FLT3 at the third parity, and rs426897754 of NLRP5 at the second and third parity can significantly increase the lambing rate of Small-tailed Han sheep (p<0.05). Moreover, bioinformatic analysis indicated changes in secondary and tertiary protein structures after g.37866222C > T mutation in TGIF1. With respect to the molecular mechanisms, further verification is required that FLT3 rs421947730 and NLRP5 rs426897754 interacting with important proteins could increase the lambing rate. Our study suggests that TGIF1, FLT3, and NLRP5 might serve as valuable genetic markers affecting the LS in sheep.
All experimental procedures involving sheep were supported by the Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China). Ethics approval was offered by the animal ethics committee of IAS-CAAS (no. IAS2020-63).
Data availability is not applicable to this article, as no new data were created or analyzed in this study.
MC, XW, and RD designed the experiments, and LT and XH carried them out. SC prepared the paper with contributions from all co-authors.
The contact author has declared that neither they nor their co-authors have any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research was funded by the National Natural Science Foundation of China (grant no. 31772580), the China Agriculture Research System of MOF and MARA (grant no. CARS-38), and the Agricultural Science and Technology Innovation Program of China (grant nos. CAAS-ZDRW202106 and ASTIP-IAS13).
This research has been supported by the National Natural Science Foundation of China (grant no. 31772580), the earmarked fund of the China Agriculture Research System (grant no. CARS-38), and the Agricultural Science and Technology Innovation Program (grant nos. CAAS-ZDRW202106 and ASTIP-IAS13).
This paper was edited by Antke-Elsabe Freifrau von Tiele-Winckler and reviewed by two anonymous referees.
An, X. P., Hou, J. X., Lei, Y. N., Gao, T. Y., Song, Y. X., Wang, J. G., and Cao, B. Y.: Two mutations in the 5′-flanking region of the KITLG gene are associated with litter size of dairy goats, Anim. Genet., 46, 308–311, https://doi.org/10.1111/age.12277, 2015.
Arfuso, F., Saoca, C., Fortino, G., Santulli, A., Fazio, F., Rizzo, M., and Piccione, G.: Gonad histological observation and multilocus microsatellite analysis in a sample of Mediterranean sea bass Dicentrarchus labrax (Linnaeus, 1758) intended for breeding: a preliminary study, Cah. Biol. Mar., 58, 379–386, https://doi.org/10.21411/CBM.A.F1D01D9B, 2017.
Ashkenazy. H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., and Ben-Tal, N.: ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules, Nucleic Acids Res., 44, W344–W350, https://doi.org/10.1093/nar/gkw408, 2016.
Bebbere, D., Ariu, F., Bogliolo, L., Masala, L., Murrone, O., Fattorini, M., Falchi, L., and Ledda, S.: Expression of maternally derived KHDC3, NLRP5, OOEP and TLE6 is associated with oocyte developmental competence in the ovine species, BMC Dev. Biol., 14, 1–13, https://doi.org/10.1186/s12861-014-0040-y, 2014.
Carcangiu, V., Giannetto, C., Luridiana, S., Fazio, F., Mura, M. C., Parmeggiani, A., and Piccione, G.: Seasons induce changes in the daily rhythm of plasma melatonin in goats (Capra hircus), Anim. Biol., 65, 13–20, https://doi.org/10.1163/15707563-00002456, 2015.
Chang, H., Nozawa, K., Liu, X. L., Geng, S. M., Ren, Z. J., Qin, G. Q., Li, X. G., Sun, J. M., Zheng, H. L., and Song, J. Z.: On phylogenetic relationships among native goat populations along the middle and lower Yellow River Valley, Asian-Aust, J. Anim. Sci., 13, 137–148, 2000.
Chang, H., Nozawa, K., Liu, X. L., Geng, S. M., Ren, Z. J., Qin, G. Q., Li, X. G., Sun, J. M., Zheng, H. L., and Song, J. Z.: On phylogenetic relationships among native goat populations along the middle and lower Yellow River Valley, Asian-Aust, J. Anim. Sci., 13, 137–148, 2020.
Chemineau, P., Daveau, A., Maurice, F., and Thimonier, J.: Seasonality and light control of reproduction in the goat, 38th Annual Meeting of the European Association for Animal Production, Lisbon, Portugal, 2, 906–907, 1987.
Chong, Y. Q., Huang, H. R., Liu, G. Q., Jiang, X. P., and Rong, W. H.: A single nucleotide polymorphism in the zona pellucida 3 gene is associated with the first parity litter size in Hu sheep, Anim. Reprod. Sci., 193, 26–32, https://doi.org/10.1016/j.anireprosci.2018.03.028, 2018.
Chu, M. X., Liu, Z. H., Jiao, C. L., He, Y. Q., Fang, L., Ye, S. C., Chen, G. H., and Wang, J. Y.: Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries), J. Anim. Sci., 85, 598–603, https://doi.org/10.2527/jas.2006-324, 2007.
Dekkers, J. C. M.: Commercial application of marker-and gene-assisted selection in livestock: strategies and lessons, J. Anim. Sci., 82, E313–E328, https://doi.org/10.2527/2004.8213_supplE313x, 2004.
Deveson, S. L., Forsyth, I. A., and Arendt, J.: Induced out-of-season breeding in British Saanen dairy goats: use of artificial photoperiods and/or melatonin administration, Anim. Reprod. Sci., 29, 1–15, https://doi.org/10.1016/0378-4320(92)90015-6, 1992.
Ellis, J. A. and Ong, B.: The MassARRAY® system for targeted SNP genotyping, Methods. Mol. Bio., 1492, 77–94, https://doi.org/10.1007/978-1-4939-6442-0_5, 2017.
Feary, E. S., Juengel, J. L., Smith, P., French, M. C., O'Connell, A. R., Lawrence, S. B., Galloway, S. M., Davis, G. H., and McNatty, K. P.: Patterns of expression of messenger RNAs encoding GDF9, BMP15, TGFBR1, BMPR1B, and BMPR2 during follicular development and characterization of ovarian follicular populations in ewes carrying the Woodlands FecX2 W mutation, Biol. Reprod., 77, 990–998, https://doi.org/10.1095/biolreprod.107.062752, 2007.
Fernandes, R., Tsuda, C., Perumalsamy, A. L., Naranian, T., Chong, J., Acton, B. M., Tong, Z. B., Nelson, L. M., and Jurisicova, A.: NLRP5 mediates mitochondrial function in mouse oocytes and embryos, Biol. Reprod., 86, 1–10, https://doi.org/10.1095/biolreprod.111.093583, 2012.
Giannetto, C, Carcangiu, V, Luridiana, S, Parmeggiani, A., and Piccione, G.: Twenty-four-hour rhythm patterns of plasma melatonin in short-day and long-day breeders maintained under natural environmental conditions, Chronobiol. Int., 37, 974–979, https://doi.org/10.1080/07420528.2020.1772808, 2020.
Giannetto, C., Cannella, V., Giudice, E., Guercio, A., Arfuso, F., and Piccione, G.: Clock genes determination in whole blood in goats housed under a long light cycle, Chronobiol. Int., 38, 1283–1289, https://doi.org/10.1080/07420528.2021.1928158, 2021.
Guca, E., Suñol, D., Ruiz, L., Konkol, A., Cordero, J., Torner, C., Aragon, E., Martin-Malpartida, P., Riera, A., and Macias, M. J.: TGIF1 homeodomain interacts with Smad MH1 domain and represses TGF-β signaling, Nucleic Acids Res., 46, 9220–9235, https://doi.org/10.1093/nar/gky680, 2018.
Guna, A. and Hegde, R. S.: Transmembrane domain recognition during membrane protein biogenesis and quality control, Curr. Biol., 28, R498–R511, https://doi.org/10.1016/j.cub.2018.02.004, 2018.
Hanrahan, J. P., Gregan, S. M., Mulsant, P., Mullen, M., Davis, G. H., Powell, R., and Galloway, S. M.: Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries), Biol. Reprod., 70, 900–909, https://doi.org/10.1095/biolreprod.103.023093, 2004.
He, X., Zhang, Z., Liu, Q., and Chu, M.: Polymorphisms of the melatonin receptor 1 A gene that affects the reproductive seasonality and litter size in Small Tail Han sheep, Reprod. Domest. Anim., 54, 1400–1410, https://doi.org/10.1111/rda.13538, 2019.
Hui, T. Y. J. and Burt, A.: Estimating linkage disequilibrium from genotypes under Hardy–Weinberg equilibrium, BMC Genet., 21, 1–11, https://doi.org/10.1186/s12863-020-0818-9, 2020.
Hyman, C. A., Bartholin, L., Newfeld, S. J., and Wotton, D.: Drosophila TGIF proteins are transcriptional activators, Mol. Cell. Biol., 23, 9262–9274, https://doi.org/10.1128/mcb.23.24.9262-9274.2003, 2003.
Jiang, Y., Xie, M., Chen, W., Talbot, R., Maddox, J. F., Faraut, T., Wu, C., Muzny, D. M,, Li, Y., Zhang, W., Stanton, J. A., Brauning, R., Barris, W. C., Hourlier, T., Aken, B. L., Searle, S. M. J., Adelson, D. L., Bian, C., Cam, G. R., Chen, Y., Cheng, S., DeSilva, U., Dixen, K., Dong, Y., Fan, G., Franklin, I. R., Fu, S., Guan, R., Highland, M. A., Holder, M. E., Huang, G., Ingham, A. B., Jhangiani, S. N., Kalra, D., Kovar, C. L., Lee, S. L., Liu, W., Liu, X., Lu, C., Lv, T., Mathew, T., McWilliam, S., Menzies, M., Pan, S., Robelin, D., Servin, B., Townley, D., Wang, W., Wei, B., White, S. N., Yang, X., Ye, C., Yue, Y., Zeng, P., Zhou, Q., Hansen, J. B., Kristensen, K., Gibbs, R. A., Flicek, P., Warkup, C. C., Jones, H. E., Oddy, V. H., Nicholas, F. W., McEwan, J. C., Kijas, J., Wang, J., Worley, K. C., Archibald, A. L., Cockett, N., Xu, X., Wang, W., and Dalrymple, B. P.: The sheep genome illuminates biology of the rumen and lipid metabolism, Science, 344, 1168–1173, https://doi.org/10.1126/science.1252806, 2014.
Joyce, I. M., Clark, A. T., Pendola, F. L., and Eppig, J. J.: Comparison of recombinant growth differentiation factor-9 and oocyte regulation of KIT ligand messenger ribonucleic acid expression in mouse ovarian follicles, Biol. Reprod., 63, 1669–1675, https://doi.org/10.1095/biolreprod63.6.1669, 2000.
Lyman, S. D. and Jacobsen, S. E. W.: c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities, Blood, 91, 1101–1134, https://doi.org/10.1182/blood.V91.4.1101, 1998.
Mahdavi, M., Nanekarani, S., and Hosseini, S. D.: Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep, Anim. Reprod. Sci., 147, 93–98, https://doi.org/10.1016/j.anireprosci.2014.04.003, 2014.
Mistry, D. S., Tsutsumi, R., Fernandez, M., Sharma, S., Cardenas, S. A., Lawson, M. A., and Webster, N. J. G.: Gonadotropin-releasing hormone pulse sensitivity of follicle-stimulating hormone-β gene is mediated by differential expression of positive regulatory activator protein 1 factors and corepressors SKIL and TGIF1, Mol. Endocrinol., 25, 1387–1403, https://doi.org/10.1210/me.2011-0032, 2011.
Molik, E., Misztal, T., Romanowicz, K., and Zieba, D.: Short-day and melatonin effects on milking parameters, prolactin profiles and growth-hormone secretion in lactating sheep, Small Ruminant Res., 109, 182–187, https://doi.org/10.1016/j.smallrumres.2012.10.006, 2013.
Mura, M. C., Luridiana, S., Vacca, G. M., Bini, P. P., and Carcangiu, V.: Effect of genotype at the MTNR1 A locus and melatonin treatment on first conception in Sarda ewe lambs, Theriogenology, 74, 1579–1586, https://doi.org/10.1016/j.theriogenology.2010.06.028, 2010.
Nishijima, C., Kawamura, K., Okamoto, N., Sato, Y., Kawamura, N., Ishizuka, B., Tanaka, M., and Suzuki, N.: Regulation of preimplantation embryo development in mice by FMS-like tyrosine kinase 3 ligand, Journal of Mammalian Ova Research, 31, 45–51, https://doi.org/10.1274/jmor.31.45, 2014.
Notter, D. R.: Genetic improvement of reproductive efficiency of sheep and goats, Anim. Reprod. Sci., 130, 147–151, https://doi.org/10.1016/j.anireprosci.2012.01.008, 2012.
Ohsugi, M., Zheng, P., Baibakov, B., Li, L., and Dean, J.: Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos, Development, 135, 259–269, https://doi.org/10.1242/dev.011445, 2008.
Okamura, Y., Myoumoto, A., Manabe, N., Tanaka, N., Okamura, H., and Fukumoto, M.: Protein tyrosine kinase expression in the porcine ovary, Mol. Hum. Reprod., 7, 723–729, https://doi.org/10.1093/molehr/7.8.723, 2001.
Souza, C. J. H., MacDougall, C., Campbell, B. K., McNeilly, A. S., and Baird, D. T.: The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene, J. Endocrinol., 169, R1, https://doi.org/10.1677/joe.0.169r001, 2001.
Takahashi, S.: Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications, J. Hematol. Oncol., 4, 13, https://doi.org/10.1186/1756-8722-4-13, 2011.
Takahashi, S., Harigae, H., Kaku, M., Sasaki, T., and Licht, J. D.: Flt3 mutation activates p21WAF1/CIP1 gene expression through the action of STAT5, Biochem. Bioph. Res. Co., 316, 85–92, https://doi.org/10.1016/j.bbrc.2004.02.018, 2004.
Tong, B., Wang, J., Cheng, Z., Liu, J., Wu, Y., Li, Y., Bai, C., Zhao, S., Yu, H., and Li, G.: Novel variants in GDF9 gene affect promoter activity and litter size in Mongolia sheep, Genes, 11, 375, https://doi.org/10.3390/genes11040375, 2020.
Tong, Z. B., Gold, L., De Pol, A., Vanevski, K., Dorward, H., Sena, P., Palumbo, C., Bondy, C. A., and Nelson, L. M.: Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development, Endocrinology, 145, 1427–1434, https://doi.org/10.1210/en.2003-1160, 2004.
Velummailum, R.: Involvement of Nlrp5 in the Maintenance of Genome Integrity in Murine Oocytes, MSc thesis, University of Toronto, Canada, 135 pp., 2011.
Wang, W., La, Y. F., Zhou, X., Zhang, X. X., Li, F. D., and Liu, B.: The genetic polymorphisms of TGFβ superfamily genes are associated with litter size in a Chinese indigenous sheep breed (Hu sheep), Anim. Reprod. Sci., 189, 19–29, https://doi.org/10.1016/j.anireprosci.2017.12.003, 2018.
Williams, J.: The use of marker-assisted selection in animal breeding and biotechnology, Rev. Sci. Tech. Oie, 24, 379–391, https://doi.org/10.20506/rst.24.1.1571, 2005.
Yoshida, H., Takakura, N., Kataoka, H., Kunisada, T., Okamura, H., and Nishikawa, S.: Stepwise requirement ofc-kit tyrosine kinase in mouse ovarian follicle development, Dev. Biol., 184, 122–137, https://doi.org/10.1006/dbio.1997.8503, 1997.
Zhang, Z., Liu, Q., Di, R., Hu, W., Wang, X., He, X., Ma, L., and Chu, M.: Single nucleotide polymorphisms in BMP2 and BMP7 and the association with litter size in Small Tail Han sheep, Anim. Reprod. Sci., 204, 183–192, https://doi.org/10.1016/j.anireprosci.2019.04.001, 2019.
Zhang, Z., He, X., Liu, Q., Tang, J., Di, R., and Chu, M.: TGIF1 and SF1 polymorphisms are associated with litter size in Small Tail Han sheep, Reprod. Domest. Anim., 55, 1145–1153, https://doi.org/10.1111/rda.13753, 2020.