the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Determination of genetic variation within the DYRK2 gene and its associations with milk traits in cattle
Cui Mao
Xing Ju
Haijian Cheng
Xixia Huang
Fugui Jiang
Yuni Yao
Xianyong Lan
Enliang Song
To speed up the progress of marker-assisted selection (MAS) in cattle breeding, the dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2), cadherin 2 (CDH2), and kinesin family member 1A (KIF1A) genes were chosen based on our pervious genome-wide association study (GWAS) analysis results. DYRK2 is a kinase that may participate in cell growth and/or development; it shows phosphorylation activity toward serine, threonine, and tyrosine fragments of proteins, and it is different from other protein kinases. The CDH2 gene encodes a classic cadherin, which is a member of the cadherin superfamily. The protein encoded by KIF1A is a member of the kinesin family and plays a role in the transportation of membrane organelles along axon microtubules. We detected insertion/deletion (InDel) variation in these three candidate genes in 438 individual cattle (Xinjiang Brown cattle and Wagyu × Luxi crossbreed cattle). Only DYRK2-P3-11 bp was polymorphic and genotyped. The polymorphism information content of DYRK2-P3-11 bp was 0.336. Correlation analyses showed that InDel polymorphism was significantly associated with six different milk traits. These findings may aid future analyses of InDel genotypes in cattle breeds, and speed up the progress of MAS in cattle breeding.
- Article
(416 KB) - Full-text XML
- BibTeX
- EndNote
Cattle are economically significant livestock. In China, Xinjiang Brown cattle (XJBC) and Wagyu × Luxi cross cattle (WLC) are high-quality domestic breeds. XJBC are an improved breed of local Chinese yellow cattle, which is obtained by crossing Chinese yellow with Brown Swiss cattle, Alatuowu cattle, and Kostroma cattle (Lin et al., 2010; Li et al., 2018). WLC are a new high-grade beef breed obtained by using modern biotechnology combined with conventional breeding techniques after 10 years of relentless effort (Wang et al., 2015). However, data on the association between their genetic background and milk traits are limited.
Milk traits are an important phenotype of cattle (Andersson et al., 2001). To have better milk production, genetic variation between cattle varieties and between major genes could be appropriately used to establish breeding programs (K. Wang et al., 2018). Genome-wide sequencing and association studies (GWASs) have been used to research genetic variation correlated with milk traits. However, many of the potential genes affecting milk traits have not been fully confirmed (Lai et al., 2016; Mota et al., 2017). To solve this problem, GWAS analyses have been used to screen large livestock populations.
Based on previous GWASs on XJBC (Zhou et al., 2019), we analyzed dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2), cadherin 2 (CDH2), and kinesin family member 1A (KIF1A) genes. DYRK2 is a kinase that may participate in cell growth and/or development. It shows phosphorylation activity toward serine, threonine, and tyrosine fragments of proteins, and it is different from other protein kinases. As a novel phosphorylation-regulated kinase, DYRK2 induces the phosphorylation of c-Myc in cancer cells and controls p53 by phosphorylation at Ser46 in response to DNA damage (Maddika et al., 2009; Taira et al., 2012). It plays a role in breast development (Tolleson et al., 2017). The CDH2 gene encodes a classic cadherin, a member of the cadherin superfamily (Kumari et al., 2018). The protein encoded by KIF1A is a member of the kinesin family, and it plays a role in the transportation of membrane organelles along axon microtubules (Lee et al., 2003).
XJBC and WLC represent important scientific achievements in China; therefore, it is important to further develop and promote these two breeds. However, traditional breeding methods are costly. Genetic molecular markers such as insertion/deletion (InDel) variation, copy number variation (CNV), and single-nucleotide polymorphisms (SNP) are widely used to assess gene function (Yang et al., 2017; Chen et al., 2019). InDel markers, in particular, are inexpensive, a time-saver, and convenient (Jander et al., 2002; Yang et al., 2017; Ju et al., 2020). Many studies have reported InDels in several genes in cattle (Meo et al., 2005), however, InDel variation in DYRK2, CHD2, and KIF1A in XJBC and WLC have not been detected. Therefore, in this study, 438 individuals representing these two cattle breeds were collected to detect and evaluate InDel variation and any correlations with milk traits. Our findings may aid the future assessment and use of genetic variants associated with phenotypic traits to improve cattle farming.
2.1 Ethics statement
All experimental animals were raised and used in accordance with local policies and animal welfare laws, and all the experimental procedures were approved by the Faculty Animal Policy and Welfare Committee of Shandong Academy of Agricultural Sciences (FAPWC-SDAAS) and Xinjiang Agricultural University (FAPWC-XJAU).
2.2 Animal samples and genomic DNA collection
In all, 438 individuals of the XJBC (n=388, Xinjiang Province) and WLC (n=50, Shandong Province) cattle breeds were obtained. The following phenotypic characteristics were recorded for each breed: 305 d milk yield (305M), milk fat percentage (FP), milk protein percentage (PP), milk fat yield (FY), milk protein yield (PY), and somatic cell score (SCS) (Peng et al., 2019). DNA samples were extracted from white blood cells using phenolic chloroform extraction (frozen at −80 ∘C) (Zhang et al., 2015). After measurement of DNA using a Nanodrop 1000 spectrophotometer (Waltham, MA, USA), the concentrations of all DNA samples were diluted to 50 ng µL−1 and temporarily stored at 4 ∘C (Lan et al., 2013).
2.3 InDel loci detection and DNA sequencing
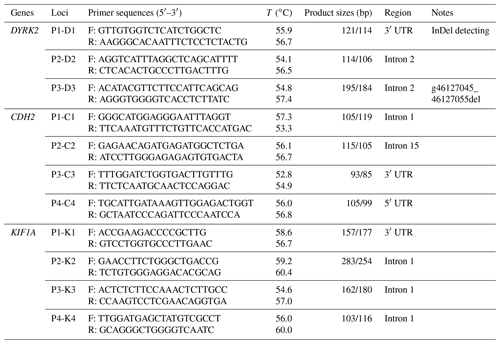
According to the whole sequence of bovine DYRK2 (GenBank no. NC_037332.1), CDH2 (GenBank no. NC_037351.1), KIF1A (GenBank no. NC_037330.1) genes, a total of 11 potential InDel variants located in 5′ untranslated regions, 3′ untranslated regions, and introns were found in the Ensembl database (https://asia.ensembl.org/index.html, last access: 15 April 2019). Relevant primers were designed using Primer Premier software 5.0 (Premier Biosoft International, Palo Alto, CA, USA) to reference the bovine CDH2 gene sequence, bovine DYRK2 gene sequence, and bovine KIF1A gene sequence (Table 1).
Polymerase chain reaction (PCR) amplification was performed as previously reported (Jin et al., 2016; Li et al., 2017; Xu et al., 2018; Ju et al., 2020), and products were separated on 3.5 % agarose gels. PCR products were sequenced when each primer pair and had different genotypes (Yang et al., 2016).
2.4 Amplification and genotyping
According to the mutation frequency and sample size of InDel loci, we used the pooling method to determine 11 pairs of primers and identified one locus (DYRK2-P3-11 bp) in XJBC (Table 1). PCR was carried out in a total reaction mixture of 13 µL, consisting of 6.5 µL 2× MIX (i.e. 2× PCR Taq MasterMix) (TsingKe, Xi'an, China), 0.3 µL each primer (forward and reverse primers), 0.3 µL genomic DNA, and 5.6 µL ddH2O (double-distilled water).
We used the Touchdown PCR program as follows: initial denaturation for 4 min at 95 ∘C, denaturation for 30 s at 94 ∘C, cycling 18 times; an annealing step for 30 s at 68 ∘C (1 ∘C reduction per cycle) with extension for 1000 bp min−1 at 72 ∘C; another 30 cycles (Czarnik et al., 2009; K. Wang et al., 2020); cooling to 4 ∘C; and detection of PCR products by 1.5 % agarose gel electrophoresis.
2.5 Statistical analyses
We used the chi-square test (χ2) to determine whether SNP variation was in Hardy–Weinberg equilibrium (HWE) (Pan et al., 2013). Furthermore, we used SPSS software (version 18.0) to analyze variance (ANOVA) and the independent sample T test to explore the correlation between InDel loci and milk traits (e.g. 305M (kg)). When necessary, we performed Bonferroni corrections for multiple comparisons (X. Y. Wang et al., 2018). Non-parametric (Kruskal–Wallis) tests in SPSS (version 18.0) were used to analyze data when homogeneity of variances did not follow a normal distribution. P<0.05 was considered statistically significant. In addition, correlations between these traits and polymorphic loci were analyzed (Peng et al., 2019).
Finally, association analyses between milk traits and InDel loci were carried out. The general linear model was as follows:
where Yij is the phenotypic value of milk traits, μ is the overall population mean, Pj is the fixed effect of parity, Gi is the fixed effect of genotype, and eijk is the random error.
3.1 Identification of DYRK2, CDH2, and KIF1A gene InDel polymorphisms
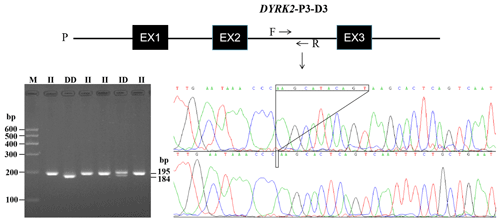
One InDel locus in DYRK2 was identified in XJBC, and it had different genotypes (II, ID, and DD) (Fig. 1). Specifically, there was an 11 bp deletion at intron 2 (DYRK2-P3-11 bp) (Table 1). Every InDel polymorphism had two or three distinct genotypes: a longer DNA fragment indicating genotype insertion–insertion (II), a shorter DNA fragment indicating genotype deletion–deletion (DD), and two or three (homoduplex) bands indicating genotype insertion–deletion (ID). The sequence diagrams for this InDel are shown in Fig. 1.
3.2 Genetic parameters calculation
We listed the frequency of population parameters for this InDel and found that the I allele (0.690) of DYRK2-P3-11 bp was more frequent than the D (0.310) allele (Table 2). Parameter analyses showed that the PIC for this InDel was 0.336, indicating moderate genetic diversity ().
Table 2Frequencies of genotypes and alleles and diversity parameter for InDel of DYRK2 gene in cattle.
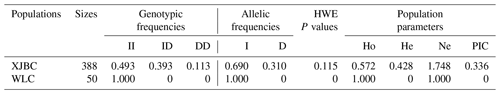
Note that HWE represents Hardy–Weinberg equilibrium, Ho represents homozygosity, He represents heterozygosity, Ne represents effective allele number, and PIC represents polymorphism information content.
3.3 Novel InDel polymorphisms had significant relationships with milk traits
Next, we studied the correlation between this XJBC DYRK2 InDel and milk traits. The InDel locus showed a significant relationship with milk traits (Table 3), such as 305M (P<0.05), PY (P<0.05) in the fourth parity, and FY (P<0.05) in the sixth parity.
Table 3Relationship between InDel polymorphism of DYRK2-P3-D3 locus and milk traits of XJBC.
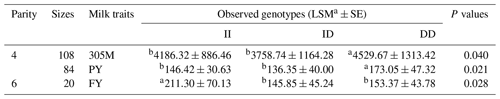
Note that 305M represents 305 d milk yield, FY represents fat yield, SCS represents somatic cell score, FP represents fat percent, PY represents protein yield, and PP represents protein percent. The values with different letters (a and b) within the same row are significant at P<0.05 and P<0.00, respectively.
In recent years, InDel makers have been widely used in MAS-based animal breeding (Wang et al., 2019; Z. Wang et al., 2020). InDel mutations of some candidate genes can be used to select excellent phenotypic traits of cattle (Cui et al., 2018; Han et al., 2018). In a previous study, we found that InDel variation in some candidate genes can be used to choose better phenotypic traits in cattle (Wang et al., 2017; Kang et al., 2019). We also found that DYRK2 was closely associated with milk traits in XJBC (Zhou et al., 2019).
DYRK2 is a part of the CMGC group of protein kinases. It contains a conservative kinase domain structure and a neighboring N-terminal DYRK homology (DH) box. Regulation irregularities in this gene may be related to the occurrence of human breast cancer (Mimoto et al., 2013; Enomoto et al., 2014). This gene has been confirmed to be involved in the development of bovine mammary glands (Tolleson et al., 2017). DYRK2 is located on chromosome 5 (BTA5), and the 11 bp InDel that we identified was significantly correlated with milk traits in XJBC.

Figure 2Bioinformatics predict transcription factor binding sites on the bovine DYRK2 gene 11 bp InDel sequences.
There have been some studies on DYRK2, but InDels are often neglected, particularly in introns and UTR (untranslated region) mutations (Maddika et al., 2009; Taira et al., 2012; Yang et al., 2016; Ju et al., 2020). Here, we identified InDel in introns, and there were insertion and deletion mutations as well (Hu et al., 2011; Liu et al., 2015). To the best of our knowledge, this is the first identification of a candidate gene, DYRK2, associated with milk traits in XJBC. The PIC value of DYRK2 was less than 0.4, indicating low genetic diversity. In addition, the InDel variant genotypes and allele distributions in XJBC and WLC were significantly different (P<0.05 or P<0.01) due to their genetic background. XJBC live in Xinjiang Province (northwestern China) (Lin et al., 2010; Li et al., 2018), and WLC live in Shandong Province (eastern China) (Wang et al., 2015). In addition, the genetic diversity of these cattle may differ because they have been subjected to different long-term artificial selection processes.
We found a significant correlation between the identified InDel and milk traits in XJBC, including 305M (P=0.040), PY (P=0.021) in the fourth parity, and FY (P=0.028) in the sixth parity. Correlation analyses confirmed these results. The minor-allele frequency (MAF) value of DYRK2-P3-11 bp was low (0.310). In addition, the InDel showed moderate polymorphism (PIC value ≥ 0.3) in both cattle; therefore, DYRK2-P3-11 bp (PIC value = 0.336) should be further studied.
According to previous research, intron mutations may affect the host genes factors and interaction between transcription (Van et al., 2003; Fushan et al., 2009). Therefore, we used the online software Jaspar Genereg (http://jaspar.genereg.net/matrix/MA0106.3/, last access: 10 July 2019) to predict transcription factor binding sites in the 11 bp InDel sequence. Bioinformatics analyses showed that forkhead box C1 (FOXC1) and forkhead box L1 (FOXL1), as transcription factors, could bind to the sequence in the absence of the 11 bp nucleotides (Fig. 2). This highlights a possibility that FOXC1 and FOXL1 influence milk traits in cattle. At the same time, FOXC1 and FOXL1 encode members of the forkhead and/or winged-helix box (FOX) family of transcription factors, and FOX transcription factor has a unique DNA-binding forkhead domain that plays a key role in regulating multiple processes including gene expression (Masuko et al., 2004) and cell proliferation. However, the effects of our identified InDel on FOXC1 and FOXL1 factor-induced milk traits have not been determined, and further studies are needed.
Briefly, an InDel in the DYRK2 candidate gene was significantly correlated with milk traits in XJBC. Our findings enrich the data on the genetic diversity of DYRK2 genes in cattle. Potentially useful DNA markers can be identified and used for MAS-based cattle breeding.
| CDH2 | cadherin 2 |
| DYRK2 | dual-specificity tyrosine phosphorylation regulated kinase 2 |
| KIF1A | kinesin family member 1A |
| GWASs | genome-wide association studies |
| InDel | insertion/deletion |
| MAS | mark-assisted selection |
| XJBC | Xinjiang Brown cattle |
| WLC | Wagyu × Luxi crossbreed cattle |
| 305M | 305 d milk yield |
| CNV | copy number variation |
| FP | milk fat percentage |
| FY | milk fat yield |
| PP | milk protein percentage |
| PY | milk protein yield |
| SCS | somatic cell score |
| SNP | single-nucleotide polymorphism |
| HWE | Hardy–Weinberg equilibrium |
| Ho | homozygosity |
| He | heterozygosity |
| Ne | effective allele numbers |
| PIC | polymorphism information content |
| UTR | untranslated region |
| LSM | least square estimate |
The data from this study can be accessed from the authors upon a reasonable request.
Conception and design of the study was done by CM, XJ, YY, XL, and ES. Acquisition of data was done by XJ and HC. Analysis and/or interpretation of data was done by CM and XJ. Drafting the article was done by CM, XJ, and XL. The critical review and revision was done by XL, FJ, and XH.
The authors declare that they have no conflict of interest.
We thank the Urumqi Xinjiang Brown Cattle Breeding Farm, Tacheng Area Xinjiang Brown Cattle Breeding Farm, Yili Xinhe Xinjiang Brown Cattle Breeding Farm, and the Xinjiang Tianshan Animal Husbandry for their cooperation and support. We also thank Shandong Academy of Agricultural Sciences for providing samples and phenotypic trait data of Shandong Black cattle, Shaanxi Key Laboratory of Molecular Biology for Agriculture, and the Life Science Research Core Services (LSRCS) of Northwest A&F University (North Campus) for their cooperation and support.
This research has been supported by the China Agriculture Research System (grant no. CARS-37), the “Science and technology innovation team and innovation project of ten agricultural characteristic industries in Jinan” (grant no. 201901), and the Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (grant no. CXGC2018E10).
This paper was edited by Steffen Maak and reviewed by three anonymous referees.
Andersson, L.: Genetic dissection of phenotypic diversity in farm animals, Nat. Rev. Genet., 2, 130–138, https://doi.org/10.1038/35052563, 2001.
Chen, M. Y., Yan, H. L., Wang, K., Cui, Y., Chen, R., Liu, J. W., Zhu, H. J., Qu, L., and Pan, C. Y.: Goat SPEF2: Expression profile, indel variants identification and association analysis with litter size, Theriogenology, 139, 147–155, https://doi.org/10.1016/j.theriogenology.2019.08.007, 2019.
Cui, Y., Yan, H., Wang, K., Xu, H., Zhang, X. L., Zhu, H. J., Liu, J. W., Qu, L., Lan, X. Y., and Pan, C. Y.: Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat, Front. Genet., 9, 91, https://doi.org/10.3389/fgene.2018.00091, 2018.
Czarnik, U., Grzybowski, G., Zabolewicz, T., Strychalski, J., and Kaminski, S.: Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish White-backed cattle and European bison, Genetika, 45, 453–459, https://doi.org/10.1134/s1022795409040103, 2009.
Enomoto, Y., Yamashita, Y., Yoshinaga, Y., Fukami, S., Miyahara, K., Nabeshima, K., and Iwasaki, A.: Downregulation of DYRK2 can be a predictor of recurrence in early stage breast cancer, Tumour. Biol., 35, 11021–11025, https://doi.org/10.1007/s13277-014-2413-z, 2014.
Fushan, A. A., Simons, C. T., Slack, J. P., Manichaikul, A., and Drayna, D.: Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose, Curr. Biol., 19, 1288–1293, https://doi.org/10.1016/j.cub.2009.06.015, 2009.
Han, J., Zhang, L., Wang, P., Zhang, L., Wang, P., Yang, G., Wang, S., Li, Y., and Pan, K. H.: Heterogeneity of intron presence/absence in Olifantiella sp. (Bacillariophyta) contributes to the understanding of intron loss, J. Phycol., 54, 105–113, https://doi.org/10.1111/jpy.12605, 2018.
Hu, H. F., Zhang, H., Liu, Z. L., Sun, J., Yang, Y. P., Wang, Y., Tan, G. P., and Xiang, Y. B.: Linkage disequilibrium and haplotype analysis of CD14 gene single nucleotide polymorphisms in Xinjiang Uygurs and Hans, J. Xinjiang. Medical. University., 34, 682–686, https://doi.org/10.3969/j.issn.1009-5551.2011.07.005, 2011 (in Chinese).
Jander, G., Norris, S. R., Rounsley, S. D., Bush, D. F., Levin, I. M., and Last, R. L.: Arabidopsis map-based cloning in the post-genome era, Plant. Physiol., 129, 440–450, https://doi.org/10.1104/pp.003533, 2002.
Jin, Y., Cai, H., Liu, J., Lin, F., Qi, X., Bai, Y., Lei, C., Chen, H., and Lan, X.: The 10 bp duplication insertion/deletion in the promoter region within paired box 7 gene is associated with growth traits in cattle, Arch. Anim. Breed., 59, 469–476, https://doi.org/10.5194/aab-59-469-2016, 2016.
Ju, X., Huang, X. X., Zhang, M. H., Wang, D., Wei, C., Jiang, H., and Lan, X. Y.: Effects of eight InDel variants in FHIT on milk traits in Xinjiang brown cattle, Anim. Biotechnol., https://doi.org/10.1080/10495398.2020.1724124, online first, 2020.
Kang, Z. H., Zhang, S. H., He, L. B., Zhu, H. J., Wang, Z., Yan, H. L., Huang, Y. Z., Dang, R. H., Lei, C. Z., Chen, H., Qu, L., Lan, X. Y., and Pan, C. Y.: A 14-bp functional deletion within the CMTM2 geneis significantly associated with litter size in goat, Theriogenology, 139, 49–57, https://doi.org/10.1016/j.theriogenology.2019.07.026, 2019.
Kumari, P., Singh, S. K., and Raman, R.: A novel non-coding RNA within an intron of CDH2 and association of its SNP with non-syndromic cleft lip and palate, Gene, 658, 123–128, https://doi.org/10.1016/j.gene.2018.03.017, 2018.
Lai, F. N., Zhai, H. L., Cheng, M., Ma, J. Y., Cheng, S. F., Ge, W., Zhang, G. L., Wang, J. J., Zhang, R. Q., Wang, X., Min, L. J., Song, J. Z., and Shen, W.: Whole-genome scanning for the litter sizetrait associated genes and SNPs under selection in dairy goat (Capra hircus), Sci. Rep.-UK., 6, 38096, https://doi.org/10.1038/srep38096, 2016.
Lan, X. Y., Zhao, H. Y., Li, Z. J., Zhou, R., Pan, C. Y., Lei, C. Z., and Chen, H.: Exploring the novel genetic variant of PITX1 gene and its effect on milk performance in dairy goats, J. Integr. Agr., 12, 118–126, https://doi.org/10.1016/S2095-3119(13)60212-9, 2013.
Lee, J. R., Shin, H., Ko, J., Choi, J., Lee, H., and Kim, E.: Characterization of the movement of the kinesin motor KIF1A in living cultured neurons, J. Biol. Chem., 278, 2624–2629, https://doi.org/10.1074/jbc.M211152200, 2003.
Li, J., Zhu, X. C., Ma, L., Xu, H. W., Cao, X., Luo, R. Y., Chen, H., Sun, X. Z., Cai, Y., and Lan, X. Y.: Detection of a new 20 bp insertion/deletion (InDel) within sheep PRND gene using mathematical expectation (ME) method, Prion., 11, 143–150, https://doi.org/10.1080/19336896.2017.1300740, 2017.
Li, N., Zhang, Y., Li, H. P., Han, L., Yan, X. M., Li, H. B., Du, W., Zhang, J. S., and Yu, Q. L.: Differential expression of mRNA-miRNAs related to intramuscular fat content in the longissimus dorsi in Xinjiang brown cattle, PLoS One, 13, e0206757, https://doi.org/10.1371/journal.pone.0206757, 2018.
Lin, H. K., Zhang, Y., Zhou, Z. Y., Pan, J. C., Zhang, J. S., Li, H. B., Yan, X. M., Du, W., and Ka, D.: The principal component analysis of Xinjiang Brown cattle body measurement trait, China, Animal. Husbandry & Veterinary. Medicine., 37, 130–133, http://www.chvm.net/CN/Y2010/V37/I8/130 (last access: 8 September 2020), 2010 (in Chinese).
Liu, X., Usman, T., Wang, Y., Wang, Z., Xu, X., Wu, M., Zhang, Y., Zhang, X., Li, Q., Liu, L., Shi, W., Qin, C., Geng, F., Wang, C., Tan, R., Huang, X., Liu, A., Wu, H., Tan, S., and Yu, Y.: Polymorphisms in epigenetic and meat quality related Genes in fourteen cattle breeds and association with beef quality and carcass traits, Asian. Austral. J. Anim., 28, 467–475, https://doi.org/10.5713/ajas.13.0837, 2015.
Maddika, S. and Chen, J.: Protein kinase DYRK2 is an E3-ligase specific molecular assembler, Nat. Cell Biol., 11, 409–419, https://doi.org/10.1038/ncb1848, 2009.
Masuko, K. and Masaru, K.: Human FOX gene family (review), Int. J. Oncol., 25, 1495–1500, https://doi.org/10.3892/ijo.25.5.1495, 2004.
Meo, G. P. D., Perucatti, A., Uboldi, C., Roperto, S., Incarnato, D., Roperto, F., Williams, J., Eggen, A., Ferretti, L., and Iannuzzi, I. L.: Comparative mapping of the fragile histidine triad (FHIT) gene in cattle, river buffalo, sheep and goat by FISH and assignment to BTA22 by RH-mapping: a comparison with HSA3, Anim. Genet., 36, 363–364, https://doi.org/10.1111/j.1365-2052.2005.01319.x, 2005.
Mimoto, R., Taira, H., Takahashi, T., Yamaguchi, M., Okabe, K., Uchida, Y., Miki, Y., and Yoshida, K.: DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail, Cancer. Lett., 339, 214–225, https://doi.org/10.1016/j.canlet.2013.06.005, 2013.
Mota, R. R., Guimarães, S. E. F., Fortes, M. R. S., Hayes, B., Silva, F. F., Verardo, L. L., Kelly, M. J., Campos, C. F., Guimarães, J. D., Wenceslau, R. R., Penitente-Filho, J. M., Garcia, J. F., and Moore, S.: Genome-wide association study and annotating candidate gene networks affecting age at first calving in Nellore cattle, Anim. Breed. Genet., 134, 484–492, https://doi.org/10.1111/jbg.12299, 2017.
Pan, C. Y., Wu, C. Y., Jia, W. C., Xu, Y., Lei, C. Z., Hu, S. R., Lan, X. Y., and Chen, H.: A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle, Gene, 531, 398–402, https://doi.org/10.1016/j.gene.2013.09.002, 2013.
Peng, K., Zhang, G. L., Yu, T., Cao, Y., Yu, Y. S., Chen, H., Lei, C. Z., Lan, X. Y., and Zhao, Y. M.: Detection of Indel variations within seven candidate genes and their associations with phenotypic traits in three cattle breeds, Anim. Biotechnol., 11, 1–9, https://doi.org/10.1080/10495398.2019.1620258, 2019.
Taira, N., Mimoto, R., Kurata, M., Yamaguchi, T., Kitagawa, M., Miki, Y., and Yoshida, K.: DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells, J. Clin. Invest., 122, 859–872, https://doi.org/10.1172/JCI60818, 2012.
Tolleson, M. W., Gill, C. A., Herring, A. D., Riggs, P. K., Sawyer, J. E., Sanders, J. O., and Rileye, D. G: Association of udder traits with single nucleotide polymorphisms in crossbred Bos indicus–Bos taurus cows, J. Anim. Sci., 95, 2399–2407, https://doi.org/10.2527/jas.2017.1475, 2017.
Van, L. A. S., Minh, N., Martin, B., Carine, N., Catherine, C., Laurence, M., Alan, L., Chris, S., Nadine, B., Michael, T., Goran, A., Michel, G., and Leif, A.: A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig, Nature, 425, 832–836, https://doi.org/10.1038/nature02064, 2003.
Wang, K., Yan, H. L., Xu, H., Yang, Q., Zhang, S. H., Pan, C. Y., Chen, H., Zhu, H. J., Liu, J. W., Qu, L., and Lan, X. Y.: A novel InDel within goat, casein alpha S1, gene is significantly associated with litter size, Gene, 671, 161–169, https://doi.org/10.1016/j.gene.2018.05.119, 2018.
Wang, K., Kang, Z. H., Jiang, E. H., Yan, H. L., Zhu, H. J., Liu, J. W., Qu, L., Lan, X. Y., and Pan, C. Y.: Genetic effects of DSCAML1 identified in genome-wide association study revealing strong associations with litter size and semen quality in goat (Capra hircus), Theriogenology, 146, 20–25, https://doi.org/10.1016/j.theriogenology.2020.01.079, 2020.
Wang, X. Y., Yang, Q., Wang, K., Zhang, S. H., Pan, C. Y., Chen, H., Qu, L., Yan, H. L., and Lan, X. Y.: Novel 12-bp InDel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats, Anim. Genet., 48, 735–736, https://doi.org/10.1111/age.12617, 2017.
Wang, X. Y., Yang, Q., Wang, K., Yan, H. L., Pan, C. Y., Chen, H., Liu, J. W., Zhu, H. J., Qu, L., and Lan, X. Y.: Two strongly linked single nucleotide polymorphisms (Q320P and V397I) in GDF9 gene are associated with litter size in cashmere goats, Theriogenology, 125, 115–121, https://doi.org/10.1016/j.theriogenology.2018.10.013, 2018.
Wang, X. Y., Yang, Q., Zhang, S. H., Zhang, X. Y., Pan, C. Y., Chen, H., Zhu, H. J., and Lan, X. Y.: Genetic effects of Single Nucleotide Polymorphisms in the goat GDF9 gene on prolificacy: true or false positive?, Animals., 9, 886, https://doi.org/10.3390/ani9110886, 2019.
Wang, Y. L. and Jiang, L. L.: The Adaptive Observation of Wagyu and Luxi crossbreed cattle Introduced to Wuwei city, China. Cattle, Science, 41, 36–38, 2015 (in Chinese).
Wang, Z., Zhang, X. L., Jiang, E. H., Yan, H. L., Zhu, H. J., Chen, H., Liu, J. W., Qu, L., Pan, C. Y., and Lan, X. Y.: InDels within Caprine IGF2BP1 intron 2 and the 3′-untranslated regions are associated with goat growth traits, Anim. Genet., 51, 117–121, https://doi.org/10.1111/age.12871, 2020.
Xu, W., He, H., Zheng, L., Xu, J. W., Lei, C. Z., Zhang, G. M., Dang, R. H., Niu, H., Qi, X. L., Chen, H., and Hung, Y. Z.: Detection of 19-bp deletion within PLAG1 gene and its effect on growth traits in cattle, Gene, 675, 144–149, https://doi.org/10.1016/j.gene.2018.06.041, 2018.
Yang, Q., Zhang, S. H., Cao, X. K., Liu, L. L., Lei, C. Z., Qi, X. L., Lin, F. P., Qu, W. D., Qi, X. S., Chen, H., and Lan, X. Y.: Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14 bp insertion/deletion (InDel) within the bovine PRNP gene, Prion., 10, 409–419, https://doi.org/10.1080/19336896.2016.1211593, 2016.
Yang, Q., Yan, H. L., Li, J., Xu, H., Wang, K., Zhu, H. J., Chen, H., Qu, L., and Lan, X. Y.: A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size, Anim. Genet., 48, 499–500, https://doi.org/10.1111/age.12551, 2017.
Zhang, S. H., Sun, K., Bian, Y. N., Zhao, Q., Wang, Z., Ji, C. N., and Li, C. T.: Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China, Sci. Rep.-UK., 5, 18336, https://doi.org/10.1038/srep18336, 2015.
Zhou, J. H., Liu, L. Y., Chen, C. P. J., Zhang, M. H., Lu, X., Zhang, Z. W., Huang, X. X., and Shi, Y. G.: Genome-wide association study of milk and reproductive traits in dual-purpose Xinjiang Brown cattle, BMC Genomics, 20, 827, https://doi.org/10.1186/s12864-019-6224-x, 2019.