the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Comparative transcriptome and histomorphology analysis of testis tissues from mulard and Pekin ducks
Linli Zhang
Zhenghong Zhang
Nemat O. Keyhani
Qingwu Xin
Zhongwei Miao
Zhiming Zhu
Zhengchao Wang
Junzhi Qiu
Nenzhu Zheng
Testicular transcriptomes were analyzed to characterize the
differentially expressed genes between mulard and Pekin ducks, which will
help establish gene expression datasets to assist in further determination
of the mechanisms of genetic sterility in mulard ducks. Paraffin sections
were made to compare the developmental differences in testis tissue between
mulard and Pekin ducks. Comparative transcriptome sequencing of testis
tissues was performed, and the expression of candidate genes was verified by
quantitative reverse transcription-polymerase chain reaction (qRT-PCR). In
mulard ducks, spermatogonia and spermatocytes were arranged in a disordered
manner, and no mature sperm were observed in the testis tissue. However,
different stages of development of sperm were observed in seminiferous
tubules in the testis tissue of Pekin ducks. A total of 43.84 Gb of clean
reads were assembled into 193 535 UniGenes. Of these, 2131 transcripts
exhibited differential expression (false discover rate <0.001 and
fold change ≥2), including 997 upregulated and 1134 downregulated
transcripts in mulard ducks as compared to those in Pekin duck testis
tissues. Several upregulated genes were related to reproductive functions,
including ryanodine receptor 2 (RYR2), calmodulin (CALM), argininosuccinate
synthase and delta-1-pyrroline-5-carboxylate synthetase ALDH18A1 (P5CS).
Downregulated transcripts included the testis-specific
serine/threonine-protein kinase 3, aquaporin-7 (AQP7) and glycerol kinase
GlpK (GK). The 10 related transcripts involved in the developmental biological
process were identified by GO (Gene Ontology) annotation. The KEGG (Kyoto
Encyclopedia of Genes and Genomes) pathways indicated that peroxisome
proliferator-activated receptors (PPARs) and calcium signaling pathways were
significantly (P<0.001) associated with normal testis physiology.
The differential expression of select genes implicated in reproductive
processes was verified by qRT-PCR, which was consistent with the expression
trend of transcriptome sequencing (RNA-seq). Differentially expressed candidate genes RYR2, CALM, P5CS,
AQP7 and GK were identified by transcriptional analysis in mulard and Pekin
duck testes. These were important for the normal development of the male
duck reproductive system. These data provide a framework for the further
exploration of the molecular and genetic mechanisms of sterility in mulard
ducks.
Highlights. The mulard duck is an intergeneric sterile hybrid offspring resulting from mating between Muscovy and Pekin ducks. The transcriptomes of testis tissue from mulard and Pekin ducks were systematically characterized, and differentially expressed genes were screened, in order to gain insights into potential gonad gene expression mechanisms contributing to genetic sterility in mulard ducks.
- Article
(1099 KB) - Full-text XML
- BibTeX
- EndNote
The mulard duck is a famous local variety in China and is considered a highly prized delicacy. The duck is the intergeneric hybrid progeny of a male Muscovy duck (Cairina moschata L.) and female domestic duck (Anas platyrhynchos var. domestica). The mulard duck exhibits strong heterosis, including high resistance, a high feed reward and delicate/high meat quality compared with its parents. However, there were no significant differences between the appearance of males and females, no obvious sex differentiation and sexual behaviors, and no actual seed values able to be determined for mulard ducks, which are essentially nonreproductive and considered sterile.
However, a careful examination revealed the presence of testes in nominally male mulard ducks during the breeding process, which produced a small amount of semen, indicating that these ducks may not be completely sterile. Indeed, on occasion, individuals have been identified as displaying various aspects of normal male mating, intromission or female egg laying. These findings suggest that one reason for the nonreproductive nature of the mulard duck may be related to the regulation of gene expression and cell differentiation in their reproductive organs because distantly hybridized sterility is a very complex biological phenomenon whose genetic basis is likely regulated by a diverse range of genetic pathways. Our previous research indicated that inconsistent karyotypes from parents contributed to intergenerically hybridized sterility because of incorrect meiosis and poor gonadal development (Tan et al., 1998). At present, transcriptome sequencing (RNA-Seq) technology has been widely applied to the detection of differential gene expression and functional annotation in livestock and poultry (Chen et al., 2016; Cong et al., 2013; Liu et al., 2017; Zeng et al., 2015). This has included investigations of reproductive mechanisms of genes related to follicular development in ducks (Xu et al., 2013) and differential gene expression in the ovaries of Shan Ma ducks, which compared peak laying and late laying periods (Zhu et al., 2017), as well as the examination of differentially expressed miRNAs between laying and non-laying periods (Yu et al., 2013). However, to date, differential gene expression has not been used to examine the genetic mechanism that may contribute to the sterility traits and reproductive performance of mulard ducks.
Given the particularity of sterility in mulard ducks, the present study utilized RNA-Seq technology to compare the transcriptomes in the testes of mulard and Pekin ducks. The set of differentially expressed genes (DEGs) were screened and characterized via the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for functional annotation and analysis to identify genes and genetic pathways related to the sex dysplasia phenotype of mulard ducks. These data will provide the basis for further investigation of the abnormal differentiation mechanisms in the reproductive system of mulard ducks and provide a framework for hypothesis development that could potentially lead to insights into treating or alleviating mulard duck infertility.
2.1 Animals
The experimental animals included male mulard and Pekin ducks, which were housed for 3–6 months with standardized feeding regulations in the Laboratory Animal Center, Fujian Academy of Agricultural Sciences (Fujian, China). The experimental protocol was approved by the Institutional Animal Care and Use Committee and the Ethics Committee on Animal Experimentation, Fujian Academy of Agricultural Sciences. The ethics committee approval number is FAAS-IAHV-AEC-2017-0510.
Experimental ducks were used for collecting semen at sexual maturity (180 d old). Animals were fasted for 12 h at the age of 210 d and then immediately sacrificed for sample (dissection of the testes) collection. The animals used included mulard ducks that did not show mounting/sexual behavior and Pekin ducks that exhibited normal mounting behaviors. Dissected testes were immediately placed in liquid nitrogen and stored at −80 ∘C until further processing.
2.2 Gonadal sections of mulard ducks and Pekin ducks
Two mulard and Pekin ducks were examined, and the development of testes was observed after dissection. The testis tissues were fixed in 10 % formaldehyde solution, embedded in paraffin and stained with hematoxylin and eosin to create tissue sections.
2.3 RNA extraction and library preparation for transcriptome sequencing
Two individuals from each variety were used for transcriptome sequencing and passed the biological repeat test, which indicated that the samples used in this study exhibited good biological repeatability and reasonable sample grouping and met the requirements of transcriptome sequencing; consequently, no additional materials were added. Total RNA was isolated from the testes using the RNeasy Lipid Tissue Mini Kit (QIAGEN, Germany) following the manufacturer's protocol. Sequencing libraries were generated using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) analysis following the manufacturer's recommendations, and barcodes were added to attribute sequences to each sample. To select cDNA fragments of ∼ 150–200 bp in length, the libraries were purified using the AMPure XP system (Beckman Coulter, Beverly, USA), followed by polymerase chain reaction (PCR) amplification. Library quality was assessed using the Agilent Bioanalyzer 2100 system.
2.4 Clustering and sequencing
Clustering of the barcoded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) in accordance with the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2000 platform, and paired-end reads were generated. Transcriptome assembly was accomplished based on the left fq and right fq using Trinity to determine transcription and gene expression levels. Differential gene expression analyses of the two conditions/groups (mulard versus Pekin) were performed using the DESeq R package (1.10.1). Transcript expression levels with an adjusted P value of <0.05 determined by DESeq were designated as differentially expressed.
2.5 GO and KEGG pathway enrichment analysis enrichment analysis
GO enrichment analyses of the DEGs were implemented using the top GO R packages based on the Kolmogorov–Smirnov test. DEGs were classified and analyzed by the Clusters of Orthologous Groups and KEGG databases, and P<0.05 was used as the significance enrichment standard.
2.6 Quantitative reverse transcription-polymerase chain reaction
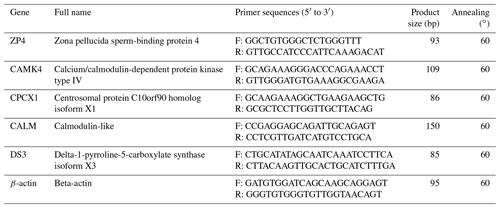
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to validate four DEGs in the cDNA pools composed of two individuals from each group. The qRT-PCR primers were designed by Primer Premier 6 and Beacon Designer 7.8 and then synthesized by Bioengineering Co., Ltd. (Shanghai). The primer sequences are listed in Table 1. The qRT-PCR was run at 95 ∘C for 1 min, followed by 40 cycles at 95 ∘C for 15 s and 63 ∘C for 25 s (fluorescence collection). The qRT-PCR was performed in a 20 L reaction mixture containing 1 L cDNA template, 10 L PowerUp SYBR® Green Master Mix (Applied Biosystems A25779), 8 µL sterile distilled water and 0.5 µL of each primer. The amount of target gene transcript relative to the internal control gene, β-actin, was calculated in accordance with the ΔΔCt method (Livak et al., 2001). Relative mRNA levels were reported as 2−ΔΔCt values. The results of three independent experiments were used for statistical analysis; P<0.05 was considered statistically significant.
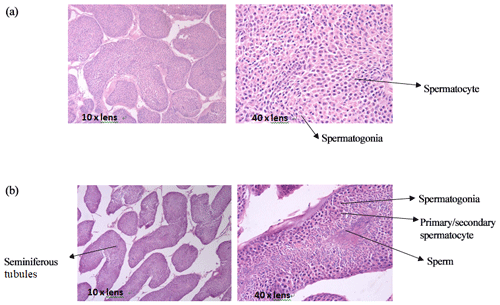
3.1 Testis tissue slices of mulard and Pekin ducks
The comparison of paraffin slices of testis tissues from mulard and Pekin ducks (Fig. 1) showed that the seminiferous tubule of the mulard duck was not obvious and had thick connective tissue. Spermatogonia and spermatocytes could be seen; however, the cells disordered, and no mature sperm were observed. The complete process of spermatogenesis was shown in the testis tissue sections of Pekin ducks. Different stages of the development of sperm were observed in the seminiferous tubules; cells close to the wall of the seminiferous tubules were oblate primitive cells, the large, round cells were primary/secondary spermatocytes, and, after meiosis, the mature haploid sperm cells were located at the center of the lumen. These reveal the differences in gonadal development of mulard and Pekin ducks on an apparent level.
3.2 Transcriptome sequencing results of testis tissues
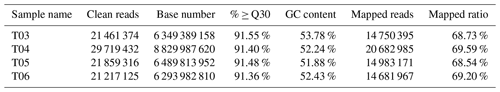
Purified mRNA libraries derived from dissected testes isolated from mulard and Pekin ducks were constructed and sequenced as detailed in Sect. 2, yielding a total of 31.05 Gb of data. After data cleanup, ∼6.34 Gb of data for each condition (∼12.68 Gb total) was obtained, and the percentage of Q30 (clean data mass of not less than 30 bases) was above 91.36 % with a GC content (the percentage of G and C bases in the total bases in clean data) of ∼52.24 % (Table 2). Additionally, the ratio of mapped reads to transcripts or the UniGene was ∼68.73 %, indicating that the sequencing quality was reliable.
3.3 Screening of DEGs and cluster analysis
FPKM (fragments per kilobase of transcript per million mapped reads) is a commonly used method to estimate gene expression levels in transcriptome data analyses, and the Pearson correlation coefficient, R, is used as an evaluation index of biological replicate correlation (Schulze et al., 2012). A correlation diagram of gene expression for each pair of biological replicate samples under the same condition was calculated (Fig. 2a), and the reliability and repeatability of the test were very high.
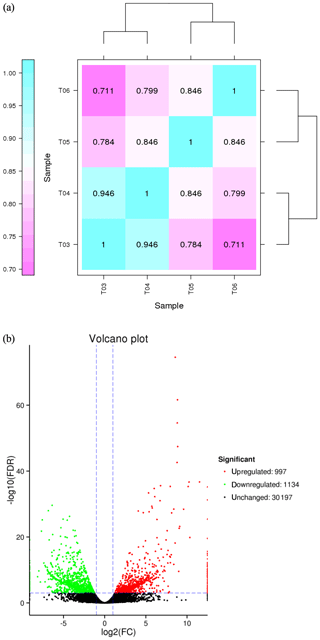
Figure 2(a) Correlation heat map of gene expression level in four samples. Notes: T03 and T04 represent two biological replicates of Pekin ducks; T05 and T06 represent two biological replicates of mulard ducks. (b) Volcano plot of differentially expressed genes The abscissa is the value of log2 (FPKM), ie, the logarithm of the mean value of expression in both samples; the ordinate is the value of log2 (FC), the logarithm of the difference in gene expression between two samples which used to measure differences in expression. Each dot represents a gene. Green dot: genes with significantly downregulated expression; red dot: genes with significantly upregulated expression; black dot: genes without differential expression. The expression level of the upregulated gene in mulard ducks is higher than that in Pekin ducks; the opposite is the case for the downregulated gene.
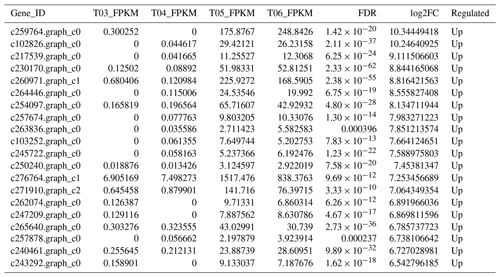
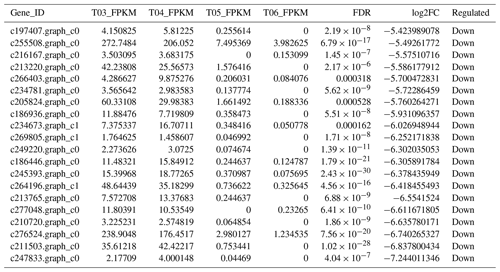
The false discovery rate (FDR) was used as a key indicator of the integrity of the DEG dataset, with 2131 transcripts identified as differentially expressed between the two duck breeds with the standard FDR < 0.001 and FC > 2.0. The DEG dataset included 997 genes that were more highly expressed in the mulard duck than in the Pekin duck and 1134 that were more highly expressed in the Pekin duck than in the mulard duck (File S1 in the Supplement). The top 20 enriched DEGs, i.e., more highly expressed, in the mulard duck, included calmodulin, CALM and DS3 (Table 3), whereas the top 20 enriched DEGs in the Pekin duck included CAMK4, CPCX1 and coiled-coil domain-containing protein 70 (Table 4). The genes more highly expressed in the mulard rather than in the Pekin duck were the regulator of G-protein signaling 2, a voltage-dependent calcium channel L type alpha-1D (CACNA1D), the ryanodine receptor 2 (RYR2), calmodulin (CALM), argininosuccinate synthase, delta-1-pyrroline-5-carboxylate synthetase ALDH18A1 (P5CS) and proline dehydrogenase (PRODH). Genes more highly expressed in the Pekin duck as compared to those of the mulard included zona pellucida sperm-binding protein 4, testis-specific serine/threonine-protein kinase 3, tyrosine 3-monooxygenase, mast/stem cell growth factor receptor Kit, Kelch-like protein 10 isoform X1, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, aryl hydrocarbon receptor, voltage-dependent calcium channel T type alpha-1I (CACNA1I), phosphatidylinositol phospholipase C zeta (PLCZ), phospholamban (PLN), phosphorylase kinase alpha/beta subunit (PHK), calcium/calmodulin-dependent protein kinase IV (CAMK4), calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterase (PDE1), creatine kinase, long-chain-fatty-acid–CoA ligase (ACSBG), aquaporin-7 (AQP7) and glycerol kinase GlpK (GK). These differences were also notable in terms of global analyses, in which the distributions of gene expression differences between the mulard and Pekin duck testis samples were distinct (Fig. 2b). Clustering analyses further demonstrated good reproducibility and reasonable grouping of the DEG dataset (Fig. 3).
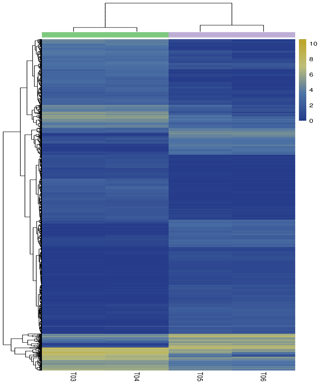
Figure 3Heatmap of the differentially expressed genes. Columns indicate individual samples, and the row represents each differentially expressed genes. The color scale represents log10 (FPKM). Notes: T03 and T04 represent two biological replicates of Pekin ducks; T05 and T06 represent two biological replicates of mulard ducks.
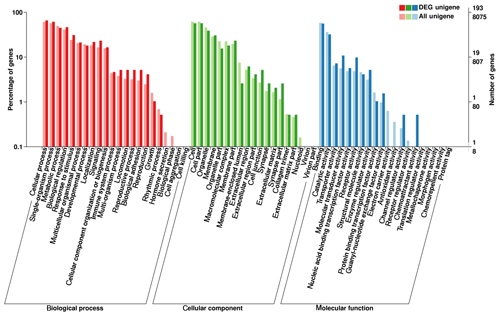
3.4 GO function enrichment
The present study used GO database analyses to compare and annotate DEGs into the three major classifications of biological process, molecular function and cellular component, resulting in the functional annotation of 786 DEGs (Fig. 4). The three major classifications were further divided into a total of 61 specific categories, consisting of 22, 19 and 20 categories, respectively (Fig. 4). The number of DEGs was most abundant in the cellular and single-organism processes within the biological process classification, in cell and cell-part categories of the cellular component classification, and in binding and catalytic activity in the molecular function classification (Fig. 4). Of note, within the biological process classification, abundant DEGs were found within categories related to reproduction and reproductive processes.
3.5 KEGG pathway enrichment analysis
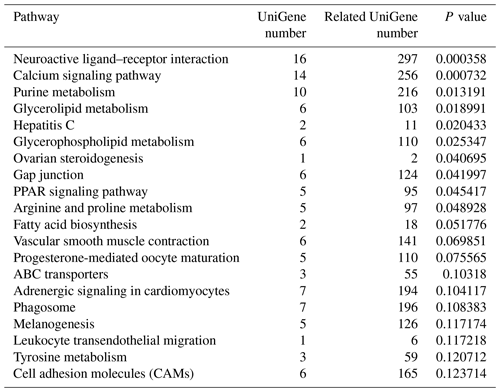
To identify the main biochemical and signaling pathways in the DEG dataset, KEGG analyses were performed, resulting in the classification of DEGs into 93 signaling pathways. The 10 pathways were significant at P<0.05, and the most significant enrichment occurred in pathways related to neuroactive ligand–receptor interactions, followed by calcium signaling pathways, purine metabolism and glycerolipid metabolism signaling pathways. Classification of the DEG dataset according to gene annotation and KEGG pathways (Fig. 5; Table 5) indicated significant enrichment in pathways related to the reproductive processes included in calcium signaling and the peroxisome proliferator-activated receptor (PPAR; nuclear hormone receptors activated by fatty acids) signaling pathways (Fig. 5). Six downregulated genes, including CACNA1I, PLCZ, PLN, PHKA_B, CAMK4 and PDE1, and three upregulated genes, CACNA1D, RYR2 and CALM, which participate in the calcium signaling pathway, were screened. Three downregulated genes, including E2.7.3.2, ACSBG and AQP7, and the upregulated gene for argininosuccinate synthase were screened in the PPAR signaling pathway. Additionally, the DEG dataset showed different degrees of enrichment particularly for those associated with signaling pathways involved in reproduction. Examples include portions of the mitogen-activated protein kinase (MAPK) signaling pathways, the gonadotropin-releasing hormone (GnRH) signaling pathway and the Wnt signaling pathways, which are used in cell–cell communication and same-cell communication and which are implicated in a range of developmental processes, including cell fate, patterning and embryonic development.
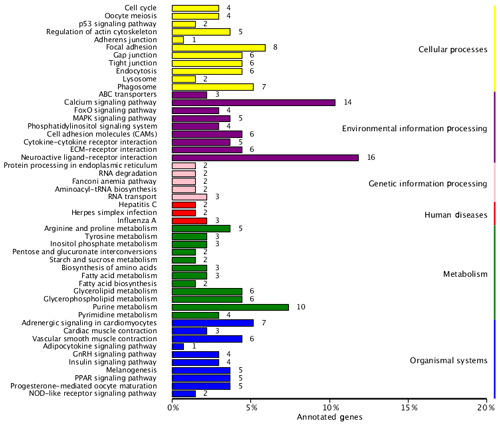
Figure 5List of KEGG pathways for differentially expressed genes. The ordinate is the name of the KEGG metabolic pathway, and the abscissa is the ratio of the number of genes annotated to pathways and the ratio of the number to the total number of genes annotated.
3.6 Verified by qRT-PCR
A set of five genes, corresponding to β-actin (CALM, DS3, ZP3, CAMK4 and CPCX1), were selected for the verification of the differential expression by qRT-PCR, as detailed in Sect. 2. Of these genes, CALM and DS3 were more highly expressed in the mulard duck compared to that of the Pekin duck, whereas the remaining three candidates were more highly expressed in the Pekin duck, as observed in the transcriptome results described above. CAMK4, CALM and ZP3 genes are the key significant differential genes related to reproduction and development, which will serve as the basis for further research on the differential genes; with FDR (false discovery rate) as the key indicator for screening differentially expressed genes, DS3 and CPCX1 are the most significantly enriched in mulard duck and Pekin duck to effectively verify the results of transcriptome sequencing. The qRT-PCR analyses of these genes using independent biological samples confirmed the transcriptomic results, with the expression levels of CALM and DS3 in mulard ducks being higher than those in Pekin ducks, whereas the expression levels of ZP3, CAMK4 and CPCX1 were higher in Pekin ducks than in mulard ducks (Fig. 6).
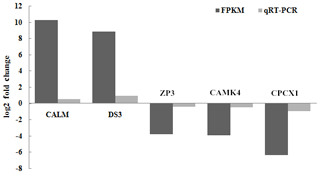
Figure 6Validation of the RNA-seq data by real-time PCR for selected genes. FPKM is a commonly used method to estimate gene expression levels in transcriptome data analysis; log2 (fold change) is the logarithm of the difference in gene expression between two samples which is used to measure differences in expression.
The present study utilized transcriptome sequencing to yield a set of DEGs comparing the testes of mulard and Pekin ducks to elucidate gene regulatory and expression pathways that might be linked to the sterile phenotype, that is to say abnormal testicular development in this study. Our analyses yielded 2131 DEGs that were analyzed by KEGG classification with significant enrichment seen in a range of biochemical metabolic and signaling pathways and with some DEGs identified as being closely associated with the reproductive process and development.
Although the development and differentiation processes leading to the mature and functional male reproductive system are complex, and our understanding is incomplete, some potentially interesting insights can be gained from our analyses. At present, various studies have shown that both p53 (Li et al., 2013) and Wnt metabolic signaling pathways (Zheng et al., 2014) contribute to male reproductive development. The MAPK extracellular signal-regulated protein kinase (MEK)/serine-threonine protein kinase (MOS)/MAPK signaling pathway has been suggested as being involved in mammalian spermatogenesis, proper meiotic (re)initiation, and subsequent aspects of mitosis and mediating cross talk to the maturation promoting factor (Almog et al., 2008; Show et al., 2008; Sun et al., 1999). Other participants identified in the mulard DEG dataset included the GnRH and its downstream signaling pathways that regulate the secretion of reproductive hormones through the gonadal axis in animals coupled with hypothalamic neurohormones that promote the release of gonadotropins from the pituitary to regulate endocrinological and developmental aspects of animal reproduction (Lee et al., 2008).
Santi et al. (1998) found that calcium (Ca2+) is an important secondary messenger in eukaryotic cells, and calcium signaling and calcium flux participate in diverse cellular processes, including spermatogenesis and the physiological activity (motility and response) of sperm. Calcium signaling also regulates phospholipase 2 (PLA2), which in turn plays an important role during the acrosome reaction that is critical for the male reproductive processes of sperm capacitation and sperm–egg fusion. The activation and regulation of sperm PLA2 activity are also mediated by specific G-protein-coupled receptors according to Yuan et al. (2003). Within this context, we identified six related genes (CACNA1I, PLCZ, PLN, PHKA/B, CAMK4 and PDE1) that were significantly more abundant in the Pekin duck than in the mulard duck, and three genes (CACNA1D, RYR2 and CALM) were more abundant in the mulard duck testis tissues than in those of the Pekin duck. Phospholipase C zeta (PLCZ) plays an important role during spermatogenesis, sperm–egg fusion, embryonic development and the catalyzation of reactions (Giesecke et al., 2010; Swann et al., 2006). Le Naour et al. (2000) showed that PLCZ activity not only affects fertilization but also plays an important role in embryonic development, particularly in promoting egg division during early embryonic development into the blastocyst stage. The loss of PLCZ expression in mulard duck testes may be a critical factor contributing to the defects related to the response to spermatogenesis, normal embryonic development, sperm–egg combination and other reproductive processes which lead to infertility in these ducks. The calcium-calmodulin-dependent protein kinase IV (CAMK4) is known to play an important role in cell fate and germ cell development. CAMK4 contributes to the regulation of meiosis, chromosome pairing and the maintenance of genome integrity during germ cell genesis and development. Therefore, the decreased relative expression of CMK4 seen in the mulard duck may result in impaired meiosis and germ cell formation. One potential mechanism by which reduced CAMK4 may act is via its regulation of protamine by phosphorylation, a reaction required for normal spermatozoal functioning (Dada et al., 2012). PPAR has pleiotropic functions including that of the process of mammalian reproduction and the regulation of spermatogenesis (Kobayashi et al., 2003). Loss of PPAR activity may lead to dysfunctions in the interstitial Leydig cells that produce testosterone in response to the luteinizing hormone leading to developmental defects in the androgen-dependent male reproductive system, including normal testicular function in adults, which negatively affects spermatogenesis and fertility. Generally, it is believed that steroid production is regulated by the trophic hormone, which promotes the transport of cholesterol from storage and synthesis sites to the mitochondrial inner membrane. Zhao et al. (2005) showed that peroxisome proliferators can block the transport induced by trophic hormones and thereby affect the male reproductive system. Our data showed that the DEGs corresponding to the PPAR pathway in the testes of mulard ducks were significantly enriched (in this instance expressed at a much lower level) as compared to those of Pekin ducks. This suggests that the lack of or decreased function of this pathway may contribute to the reproductive disorders seen in mulard ducks. This study screened and selected three downregulated genes (GK; long-chain-fatty-acid–CoA ligase, ACSBG; aquaporin-7, AQP7) and one upregulated gene (argininosuccinate synthase, AS). Among the DEGs screened in this pathway. Suzuki-Toyota et al. (1999) suggested that AQP7 is significantly expressed on the plasma membrane of sperm in the testis and epididymis. The expression of AQP7 in spermatozoa is known to affect semen quality. Low expression of the AQP7 gene in the testes of mulard ducks may greatly affect sperm production or quality, resulting in abnormal development of the testes in mulard ducks.
Some pathways that appeared abnormally expressed in the mulard duck were found that did not appear to have direct links to fertility. These included various neuroactive ligand–receptor interactions. We speculate that these may be involved in nerve body development, which may have indirect effects in terms of behavior and the activities and function of the testes. Additional pathways identified included that of purine/glycerolipid metabolism, which may function to provide nucleotide pools and signaling events critical to testis function.
A set of DEGs was identified in a comparative analysis of gene expression between testis tissues of mulard and Pekin ducks. Differential regulation of genetic pathways involved in spermatogenesis, meiosis and sperm–egg fusion was observed in mulard ducks, which provides candidate networks and hypotheses to account for the male-sterile phenotype of mulard ducks. Our data provided a foundation for further functional studies to elucidate the mechanisms of hybrid sterility, probe normal testicular development and function, and provide a means for therapeutic targeting to overcome the lack of reproductive vigor observed in the mulard duck.
The original data have been uploaded to the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA657980, accession: PRJNA657980, Fujian Academy of Agricultural Sciences, 2020).
JQ and NZ designed the experiments and LL carried them out. ZZha and ZW polished the English. NOK reviewed the paper and made comments. LZ, QX, ZM and ZZhu collected samples. LL prepared the paper with contributions from all coauthors.
The authors declare that they have no conflict of interest.
This study was supported by the National Key Research and Development Program of China (2017YFE0122000), the National Natural Science Foundation of China (U1803232, 31670026), Fujian Natural Science Foundation (2018J01041), Fujian Province Public Welfare Scientific Project (2017R1023-5), and Young Talent Innovation Foundation of Fujian Academy of Agricultural Sciences (MYQJ(S)2018-2).
This research has been supported by the National Key Research and Development Program of China (grant no. 2017YFE0122000), the National Natural Science Foundation of China (grant nos. U1803232 and 31670026), the Fujian Natural Science Foundation (grant no. 2018J01041), the Fujian Province Public Welfare Scientific Project (grant no. 2017R1023-5), and the Young Talent Innovation Foundation of Fujian Academy of Agricultural Sciences (grant no. MYQJ(S)2018-2).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Almog, T. and Naor, Z.: Mitogen activated protein kinases (MAPKs) as regulators of spermatogenesis and spermatozoa functions, Mol. Cell Endocrinol., 282, 39–44, https://doi.org/10.1016/j.mce.2007.11.011, 2008.
Chen, L., Huang X., Tian Y., Tao, Z., and Lu, L.: Identifying genes associated with blue eggshell in ducks (Anasplatyrhynchos domesticus) by transcriptome analysis, Journal of Agricultural Biotechnology, 24, 1064–1072, 2016.
Cong, F., Liu, X., Han, Z., Shao, Y., Kong, X., and Liu, S.: Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection, BMC Genomics, 14, 743, https://doi.org/10.1186/1471-2164-14-743, 2013.
Dada, R., Kumar, M., Jesudasan, R., Fernández, J. L., Gosálvez, J., and Agarwal, A.: Epigenetics and its role in male infertility, J. Assist. Reprod. Gen., 29, 213–223, 2012.
Fujian Academy of Agricultural Sciences: duck testis sequencing (Searching for differentially expressed genes in gonads of Mulard ducks and parents), NCBI, available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA657980, last access: 19 August 2020.
Giesecke, K., Sieme, H., and Distl, O.: Infertility and candidate gene markers for fertilitin stallions, Vet. J., 185, 265–271, https://doi.org/10.1016/j.tvjl.2009.07.024, 2010.
Kobayashi, T., Niimi, S., Kawanishi, T., Fukuoka, M., and Hayakawa, T.: Changes in peroxisome proliferator-activated receptor γ-regulated gene expression and inhibin/activin–follistatin system gene expression in rat testis after an administration of di-n-butyl phthalate, Toxicol. Lett., 138, 215–225, https://doi.org/10.1016/S0378-4274(02)00414-9, 2003.
Lee, V. H., Lee, L. T., and Chow, B. K.: Gonadotropin-releasing hormone: regulation of the GnRH gene, FEBS J., 275, 5458–5478, https://doi.org/10.1111/j.1742-4658.2008.06676.x, 2008.
Le Naour, F., Rubinstein, E., Jasmin, C., Prenant, M., and Boucheix, C.: Severely reduced female fertility in CD9-deficient mice, Science, 287, 319–321, 2000.
Li, G. Y., Xie, P., Li, H. Y., Hao, L., Xiong, Q., and Qiu, T.: Involment of p53, Bax, and Bcl-2 pathway in microcystins-induced apoptosis in rat testis, Environ. Toxicol., 26, 111–117, https://doi.org/10.1002/tox.20532, 2013.
Liu, Y., He, S., Zeng, T., Du, X., Shen, J., Zhao, A., and Lu, L.: Transcriptome analysis of the livers of ducklings hatched normally and with assistance, Asian Austral. J. Anim., 30, 773, https://doi.org/10.5713/ajas.16.0528, 2017.
Livak, K. J. and Schmittgen, T. D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method, Methods, 25, 402–408, https://doi.org/10.1006/meth.2001.1262, 2001.
Santi, C. M., Santos, T., Hernández-Cruz, A., and Darszon, A.: Properties of a novel pH-dependent Ca2+ permeation pathway present in male germ cells with possible roles in spermatogenesis and mature sperm function, J. Gen. Physiol., 112, 33–53, https://doi.org/10.1085/jgp.112.1.33, 1998.
Schulze, S. K., Kanwar, R., Gölzenleuchter, M., Therneau, T. M., and Beutler, A. S.: SERE: Single-parameter quality control and sample comparison for RNA-Seq, BMC Genomics, 13, 524, https://doi.org/10.1186/1471-2164-13-524, 2012.
Show, M. D., Hill, C. M., Anway, M. D., Wright, W. W., and Zirkin, B. R.: Phosphorylation of mitogen-activated protein kinase 8 (MAPK8) is associated with germ cell apoptosis and redistribution of the Bcl2-Modifying Factor (BMF), J. Androl., 29, 338–344, https://doi.org/10.2164/jandrol.107.003558, 2008.
Sun, Q. Y., Breitbart, H., and Schatten, H.: Role of the MAPK cascade in mammalian germ cells, Reprod. Fert. Develop., 11, 443–450, https://doi.org/10.1071/RD00014, 1999.
Suzuki-Toyota, F., Ishibashi, K., and Yuasa, S.: Immunohisto chemical localization of a water channel, aquaporin 7 (AQP7), in the rat testis, Cell Tissue Res., 295, 279–285, 1999.
Swann, K., Saunders, C. M., Rogers, N. T., and Lai, F. A.: PLCζ(zeta): A sperm protein tha triggers Ca2+ oscillations and egg activation in mammals, Semin. Cell Dev. Biol., 17, 264–273, https://doi.org/10.1016/j.semcdb.2006.03.009, 2006.
Tan, J., Chen, H., Song, J., and Liu, Y.: Studies on reproductive character of Mulard ducks, Journal of Fujian Agricultural Sciences, 13, 41–45, 1998.
Xu, Q., Zhao, W., Chen, Y., Tong, Y., Rong, G., Huang, Z., and Chen, G.: Transcriptome profiling of the goose (Anser cygnoides) ovaries identify laying and broodiness phenotypes, PloS one, 8, e55496, https://doi.org/10.1371/journal.pone.0055496, 2013.
Yu, D. B., Jiang, B. C., Gong, J., Dong, F. L., Lu, Y. L., Yue, H. J., Wang, Z. C., Du, W. X., and Guo, A. Y.: Identification of novel and differentially expressed microRNAs in the ovaries of laying and non-laying ducks, J. Integr. Agr., 12, 136–146, 2013.
Yuan, Y. Y., Chen, W. Y., Shi, Q. X., Mao, L. Z., Yu, S. Q., Fang, X., and Roldan, E. R. S.: Zona pellucida induces activation of phospholipase A2 during acrosomal exocytosis in guinea pig spermatozoa, Biol. Reprod., 68, 904–913, https://doi.org/10.1095/biolreprod.102.005777, 2003.
Zeng, T., Zhang, L., Li, J., Wang, D., Tian, Y., and Lu, L.: De novo assembly and characterization of muscovy duck liver transcriptome and analysis of differentially regulated genes in response to heat stress, Cell Stress and Chaperones, 20, 483–493, 2015.
Zhao, Y. and Yang, Z.: PPARs signaling pathway in mammalian reproduction, Chinese Journal of Cell Biology, 27, 14–18, 2005.
Zheng, W., Zhao, Z. S., Li, Q. F., Ban, Q., Li, H. T., and Liang, Y. W.: Analysis of difference microRNAs in female and male embryos of chicken-quail hybrid by solexa sequencing, Chinese Journal of Veterinary Science, 34, 116–122, 2014.
Zhu, Z., Miao, Z., Chen, H., Xin, Q., Li, L., Lin, R., Haung, Q., and Zheng, N.: Ovarian transcriptomic analysis of Shan Ma ducks at peak and late stages of egg production, Asian Austral. J. Anim., 30, 1215–1224, https://doi.org/10.5713/ajas.16.0470, 2017.