the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Associations of ORMDL1 gene copy number variations with growth traits in four Chinese sheep breeds
Xiaogang Wang
Xiukai Cao
Yifan Wen
Yilei Ma
Ibrahim Elsaeid Elnour
Yongzhen Huang
Xianyong Lan
Buren Chaogetu
Linyong Hu
Hong Chen
Copy number variations (CNVs) are gains and losses of genomic sequence of more than 50 bp between two individuals of a species. Also, CNV is considered to be one of the main elements affecting the phenotypic diversity and evolutionary adaptation of animals. ORMDL sphingolipid biosynthesis regulator 1 (ORMDL1) is a protein-coding gene associated with diseases and development. In our study, the polymorphism of ORMDL1 gene copy numbers in four Chinese sheep breeds (abbreviated CK, HU, STH, and LTH) was detected. In addition, we analyzed the transcriptional expression level of ORMDL1 gene in different tissues of sheep and examined the association of ORMDL1 CNV with growth traits. The statistical analysis revealed that ORMDL1 CNV was remarkably correlated with body height, heart girth, and circumference of cannon bone in HU sheep (P<0.05), and there are significant effects on body weight, body height, body length, chest depth, and height of hip cross in STH sheep (P<0.05). In conclusion, our results provide a basis for the relationship between CNV of ORMDL1 gene and sheep growth traits, suggesting that ORMDL1 CNV may be considered a promising marker for the molecular breeding of Chinese sheep.
- Article
(300 KB) - Full-text XML
-
Supplement
(140 KB) - BibTeX
- EndNote
China has a rich resource of native sheep breeds (Zhao and Li, 2017), some of which possess specific traits such as fresh and tender meat, strong stress resistance, and so on. Breeding sheep with excellent growth traits by molecular breeding technology has become an essential means of breeding. Copy number variation (CNV) might be one of the main factors affecting phenotypic diversity and evolutionary adaptation in animals, employing a wide variety of mechanisms, such as gene dosage and transcript structure alterations, to modulate organismal plasticity (Clop et al., 2012). There are four kinds of the forming mechanism of CNV, consisting of non-allelic homologous recombination (NAHR), non-homologous end-joining (NHEJ), fork stalling and template switching (FoSTeS), and L1-mediated retrotransposition (Gu et al., 2008). The recent studies showed that the different copy number types of genes in the sheep genome had different effects on the growth and development of sheep, such as several critical CNV-overlapping genes (BTG3, PTGS1, and PSPH) which were involved in fetal muscle development, prostaglandin (PG) synthesis, and bone color (Yang et al., 2018a). According to reports, the CNV map of the Chinese native sheep genome has been successfully constructed based on the Illumina Ovine SNP 600 K BeadChip array (Ma et al., 2017). Moreover, CNV detection and map construction have been accomplished in chicken (Wang et al., 2010), goat (Fontanesi et al., 2010), and cattle (Bae et al., 2010; Fadista et al., 2010).
ORMDL families consist of three genes: ORMDL1, ORMDL2, ORMDL3. ORMDL sphingolipid biosynthesis regulator 1 (ORMDL1) is a protein-coding gene which is related to sphingolipid metabolism, metabolism, ceramide homeostasis, and disease. An important paralog of this gene is ORMDL2. Studies have shown that ORMDL proteins are the primary regulators of ceramide biosynthesis and mediate feedback regulation of ceramide biosynthesis in mammalian cells (Siow and Wattenberg, 2012). ORMDLs may be involved in the regulation of ceramides during IL-1-mediated sterile inflammation (Cai et al., 2016). Studies have confirmed that polymorphisms in ORMDL genes are associated with asthma; increased ORMDL levels in asthmatic patients indicate that ORMDLs can cause asthma (Toncheva et al., 2015). In recent years, the ORMDL protein is a response to excess cholesterol, leaving the endoplasmic reticulum (ER) to activate serine palmitoyl-CoA transferase (SPT) and increase sphingomyelin biosynthesis, which buffers excess cellular cholesterol (Wang et al., 2015). Besides, the fact that human ORMDL homologs can rescue yeast mutants indicated that ORMDL is involved in protein folding in the ER (Hjelmqvist et al., 2002). We hypothesize that it may improve the function of ORMDL1 gene and affect growth traits by regulating metabolism and ceramide levels in sheep.
In this study, we found that the ORMDL1 gene has a copy number variation region (Chr2: 118 432 001–118 434 800 bp, 2800 bp), which is closely related to growth traits in sheep. Based on the Animal QTL Database, we found that the ORMDL1 CNV region is involved in a high density of quantitative trait loci (QTLs) for various traits of economic importance, especially muscle and fat development (Table 1). Moreover, there is no literature reported on the relationship between the copy number variation of ORMDL1 gene and growth traits in sheep. We analyzed the associations of ORMDL1 CNV with growth traits in four Chinese sheep breeds. The result shows that CNV of ORMDL1 gene may be used as a molecular marker for the sheep breeding.
Table 1QTLs associated with ORMDL1 CNV.
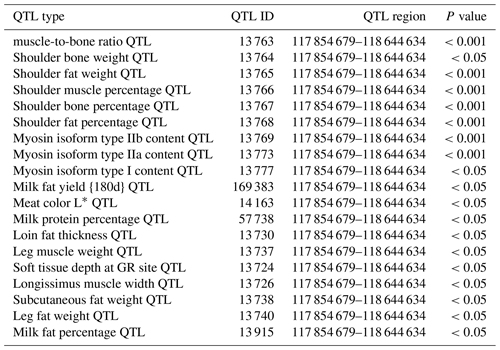
The meat color was determined using a Minolta ChromaMeter (Minolta Co., Ltd., Osaka, Japan) calibrated to a white standard using the L*, a*, b* scale. Color L* (lightness), color a* (redness), color b* (yellowness). Note that “GR” is the soft tissue depth 110 mm off the midline in the region of the 12th rib.
2.1 Animals and growth trait measurements
All experiments were approved by the Review Committee for the Use of Animal Subjects of Northwest A&F University. Four Chinese sheep breeds were tested in this study so as to detect the intergroup distribution of ORMDL1 gene copy number variations: Chaka sheep (CK, n=300, Haixi state, Qinghai province, China), Hu sheep (HU, n=198, Pingle town, Mengjin county, Henan province, China), Small-tailed Han sheep (STH, n=182, Yongjing county, Gansu province, China), and Large-tailed Han sheep (LTH, n=54, Yongjing county, Gansu province, China). As shown in Table S1 in the Supplement, the information for sex and age of four Chinese sheep breeds is provided. All 734 sheep were used to detect the frequency distributions of CNVs. All of the individuals were selected randomly as test cases. Moreover, the individuals from each breed were selected from the same breeding farm. Blood samples were obtained and genomic DNA was isolated from leukocytes with phenol–chloroform extraction. The phenotypic data of growth traits such as body weight, body length, body height, chest depth, heart girth, pipe circumference, rump width, and so on were recorded and collected for all adults in four breeds for the CNV association analysis.
2.2 Genomic DNA extraction, RNA isolation, and cDNA synthesis
All blood samples of the sheep were collected and genomic DNA was extracted from the blood samples (2 mL) by proteinase K digestion, extraction, and the phenol–chloroform methods. Tissue samples (heart, liver, spleen, lung, kidney, muscle, fat) were collected from the slaughterhouse and quickly put into liquid nitrogen. Total RNA was isolated from the different tissues with Trizol reagent (TaKaRa, Otsu, Shiga, Japan) according to the manufacturer's protocol and treated with RNase-free DNase (TaKaRa). Concentrations and purity of RNA were measured by spectrophotometry. First-strand complementary DNA (cDNA) was synthesized using a PrimeScript RT Reagent Kit (Perfect Real Time) (Clontech, TaKaRa) with 1 µg of total RNA as the template.
2.3 Primer design and amplification detection
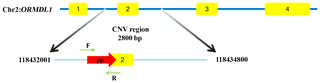
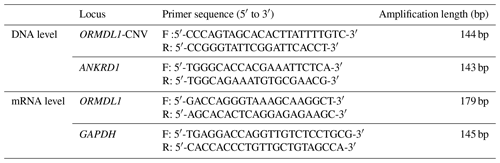
The ORMDL1 has CNV in pig and cattle according to the Animal Omics Database (http://animal.nwsuaf.edu.cn/code/index.php/main/loadByGet?address[]=main/html/panGenomeDB.php, last access: 17 September 2019). Further validation of the CNV region, including the ORMDL1 gene, was performed for sheep in this study. Our unpublished CNV data (i.e., Huang et al., unpublished data) about sheep indicated that the ORMDL1 CNV region, ORMDL1-CNV, is located in the ORMDL1 gene reference genome sequence (Oar_v4.0) from Chr2: 118 432 001–118 434 800 bp, a total of 2800 bp (Fig. 1). The primer was designed in the CNV region of ORMDL1 gene; meanwhile, the reference primer was designed using NCBI Primer-BLAST. The primer information is shown in Table 2; as illustrated in Fig. 1, the amplified fragment by primer pair (ORMDL1-CNV) was in the CNV region.
2.4 Copy number analysis of ORMDL1 gene
In this study, we researched the relative copy numbers of sheep ORMDL1 gene. Ankyrin repeat domain 1 (ANKRD1) was chosen as the internal reference gene because there is neither CNVs nor segmental duplication in the Database of Genomic Variants of ANKRD1. The copy number of ORMDL1 gene was confirmed based on the assumption that there were two copies of the DNA segment in the calibrator animals. Genomic quantitative polymerase chain reaction (qPCR) experiments were conducted using SYBR® Green in triplicate reactions. A total of 12.5 µL reaction mixtures contained 10 ng of DNA, 6.25 µL 2× RealStar Green Fast Mixture (GenStar, Beijing, China), and 10 pmol of primers. Thermal cycling conditions consisted of one cycle of 10 min at 95 ∘C followed by 40 cycles of 15 s at 95 ∘C, 60 s at 60 ∘C, and 30 s at 72 ∘C.
2.5 Expression profiling of ORMDL1 in sheep
The expression levels of ORMDL1 in different tissues of sheep were detected using qPCR on a CFX 96™ real time detection system (Bio-Rad, Hercules, CA, USA). The gene's relative expression was normalized to the expression of the sheep GAPDH gene. The qPCR experiment was performed using ORMDL1 and GAPDH gene-specific primer pairs (Table 2). The ORMDL1 gene expression levels were quantified using Gene Expression Macro software (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) by employing an optimized comparative Ct (ΔΔCt) value method, commonly designated as 2−ΔΔCt. All the experiments were repeated three times and the result was presented as mean ± SD.
2.6 Statistical analysis
In our study, the association of ORMDL1 copy numbers and mRNA expression levels were analyzed using (L. Yang et al., 2017; M. Yang et al., 2017) and 2−ΔΔCt respectively. The copy number of ORMDL1 was divided into three types: gain (copy number >2), loss (copy number <2), and median (copy number = 2). The associations of ORMDL1 CNV with growth traits in CK, HU, STH, and LTH sheep breeds were analyzed using SPSS v18.0 software (SPSS, Inc., Chicago, IL, USA) via the general linear model method. The effects on phenotypic traits were statistically analyzed using the following model: , where Yijkl is the observation of the growth traits, μ is the overall mean of each trait, Ai is the effect due to ith age, Sj is the effect due to jth sex, CNVk is the effect of jth CNV type of ORMDL1 gene, and eijkl is the random residual error.
3.1 Distribution of CNVs of ORMDL1 in four sheep breeds
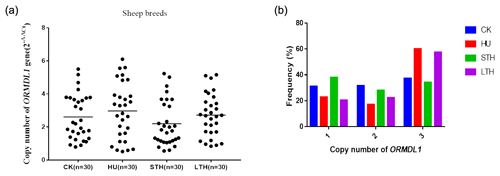
In order to investigate the distribution of ORMDL1 CNVs in different sheep breeds, we detected the copy number of ORMDL1 in CK, HU, STH, and LTH breeds. The CNV types were divided into gain (copy number >2), loss (copy number <2), and median (copy number = 2) based on the values. First, we selected 30 of each sheep breed to study whether the ORMDL1 gene has CNV. As is shown in Fig. 2a, the distribution of ORMDL1 CNV shows that the gain of copy number was more dominant than loss and median in CK, HU and LTH sheep, while the copy number loss was more dominant than gain and median in STH sheep. In addition, Fig. 2b displays the proportion of subjects with relative copy numbers of the ORMDL1 gene in the four breeds. It can be seen that the frequency of sheep (CK, HU, LTH) with a copy number of >2 was much higher than sheep with other copy numbers, especially in the HU breed (60 %). The copy numbers main varying from one copy to three copies was identified in the four breeds. All breeds show varying types of gain and loss, suggesting that the diversity of variation appeared in the ORMDL1 gene CNV region.
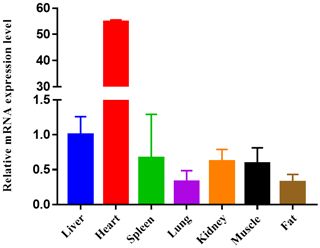
Figure 3Expression profiling of ORMDL1 gene in different tissues in sheep. The values are the averages of three independent experiments measured by 2−ΔΔCt. Error bars represent the standard deviation (SD) (n=3), the relative mRNA expression levels of ORMDL1 gene are normalized, and GAPDH was used as internal reference.
Table 3Statistical association analysis of sheep ORMDL1 gene copy number variations with growth traits in CK sheep.
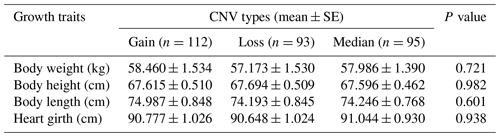
Table 4Statistical association analysis of sheep ORMDL1 gene copy number variations with growth traits in HU sheep.
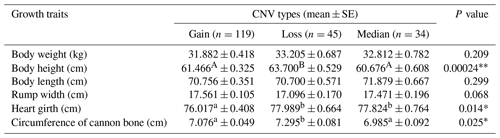
Values with different superscripts (e.g., a, b) within the same row differ significantly at * P<0.05. Values with different superscripts (e.g., A, B) within the same row differ significantly at P<0.01.
3.2 Association analysis between ORMDL1 CNV and growth traits in sheep breeds
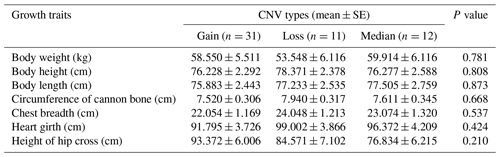
In recent years, many studies have reported that CNVs are associated with human height and livestock growth as well as growth traits (Kim et al., 2013; Zhang et al., 2018). In this study, association analysis of ORMDL1 CNVs with growth traits was performed in four sheep breeds (CK, HU, STH, LTH) by a general linear model. As is shown in Tables 3–6, no significant differences were detected among the ORMDL1 CNV types with growth traits in CK and LTH sheep (P>0.05) but there was a tendency that individuals with copy number gain had better traits than those with copy number loss or median in CK sheep. However, in the HU sheep population, there are significant effects on body height, heart girth, and circumference of cannon bone (P<0.05), especially with the body height (P=0.00024), and the individuals with copy number loss had higher values than those with gain or median; Similarly, the type of gain has better growth traits than loss and median in body weight, body height, body length, chest depth, and height of hip cross (P<0.05), especially with the body weight (P=0.001), body height (P=0.009), and height of hip cross (P=0.00). These results show that the CNV of ORMDL1 gene has a remarkable influence on sheep's growth traits (P<0.05).
Table 5Statistical association analysis of sheep ORMDL1 gene copy number variations with growth traits in STH sheep.
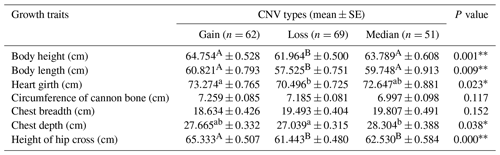
Values with different superscripts (e.g., a, b) within the same row differ significantly at * P<0.05. Values with different superscripts (e.g., A, B) within the same row differ significantly at P<0.01.
3.3 The mRNA expression level of the ORMDL1 gene in sheep tissues
To study the potential mechanism of action of CNV, here we show the mRNA expression profiling of ORMDL1 in seven different tissues (liver, heart, spleen, lung, kidney, muscle, fat) of sheep (Fig. 3). Obviously, the mRNA of ORMDL1 is widely expressed in the tissues to be tested, with the highest expression level in the heart and liver, a medium expression level in the spleen, kidney, and muscle; and relatively low expression levels in lung and fat.
Genomic polymorphisms exist in various forms including single-nucleotide variations, translocations, insertions, and CNVs (Kalman and Vitale, 2009; McKnight et al., 2010; Yang et al., 2016). Due to the broader coverage of CNV and the more considerable variation in genomic structure, more and more research on CNV has been conducted in recent years. Copy number variations have been found in human and livestock genomes with the development of whole-genome sequencing, and CNV is mainly used to study human diseases (Henrichsen et al., 2009) such as neurodevelopmental disorders (Takumi and Tamada, 2018). In animal breeding, according to reports, copy number variations can affect the economic traits of animals and lay the foundation for molecular breeding, such as in pig (Fowler et al., 2013), chicken (Lin et al., 2018), sheep (Ma et al., 2017), and cattle (Xu et al., 2014; Zheng et al., 2019). Studies have shown that a comprehensive sheep CNV map has been generated, equivalent to 6.9 % of the sheep genome (Yang et al., 2018b). However, it is necessary to use a large number of experimental individuals from various sheep breeds to identify the existence of gene CNVs and to study whether specific CNVs affect individuals. Therefore, in this study, we reported for the first time that the CNVs of the ORMDL1 gene were significantly associated with certain growth traits in the HU and STH sheep breeds.
There are many methods for the detection of copy number variation, such as SNP chip, comparative genomic hybridization (CGH) (Pinto et al., 2011), qPCR assay, and next-generation sequencing (NGS). However, qPCR is widely used as a promising means of verifying CNV due to its simple and practical qPCR assay (Xu et al., 2013). In our study, the ΔΔCt analysis and the reference gene ANKRD1 were used to confirm the selected CNV region and also to first analyze the ORMDL1 gene mRNA expression levels in different tissues of sheep. These results show that the expression level of ORMDL1 in heart and liver is higher than other tissues of sheep, suggesting that the ORMDL1 gene might play a critical role in heart development. The liver is the main organ of metabolism; therefore, we think that the effect on growth traits may be achieved through adaptive means. ORMDL1 is an important gene involved in sphingolipid metabolism, so we speculate that the ORMDL1 gene influences the sphingolipid metabolism in the liver. In addition, the ORMDL1 gene might also affect the development of muscle in sheep.
There is a difference in the distribution of ORMDL1 copy number among the four sheep breeds; this may be attributed to the diverse genetic background among breeds (Lehnert et al., 2007) or may be affected by the growing environment. The CNV region has been detected in many functional genes, and the copy number may influence the transcriptional and phenotypic variations through the dose effect. Some studies have reported that CNVs have potential effects on economic and reproductive traits in livestock (Hou et al., 2011). Additionally, some QTLs spanning the CNV fragments were related to chicken body weight, carcass weight, and breast muscle mass (Wang et al., 2012). In our study, the results show that the ORMDL1 gene significantly associated with many growth traits in HU and STH sheep, especially with body weight, body height, and heart girth. This may be due to the dose effect of the ORMDL1 gene, which in turn affects the development and deposition of muscles, leading to significant differences in body weight and heart girth.
In short, we detected and validated the copy number variations in the ORMDL1 gene in different Chinese sheep breeds. In addition, we have examined the expression of ORMDL1 gene in various tissues of sheep at the mRNA level. Correlation analysis shows that the ORMDL1 gene CNV had significant effects on the growth traits of Chinese sheep, especially in the HU and STH sheep. Therefore, our study provides elementary evidence for the functional roles of the ORMDL1 CNV in larger populations and different sheep breeds, which may provide a new idea for the potential application of CNV in sheep breeding as new molecular markers in the future.
The original data are available upon request to the corresponding author.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-62-571-2019-supplement.
HC, LH, YH, and XL designed the study. XW performed the experiments and drafted the manuscript. XC, YW, YM, and IEE helped perform the experiments and analyzed the data. BC helped collect blood samples.
The authors declare that they have no conflict of interest.
This research has been supported by the Qinghai Province major science and technology project (grant no. 2017-NK-153).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Bae, J. S., Cheong, H. S., Kim, L. H., NamGung, S., Park, T. J., Chun, J. Y., Kim, J. Y., Pasaje, C. F., Lee, J. S., and Shin, H. D.: Identification of copy number variations and common deletion polymorphisms in cattle, BMC Genomics, 11, 232, https://doi.org/10.1186/1471-2164-11-232, 2010.
Cai, L., Oyeniran, C., Biswas, D. D., Allegood, J., Milstien, S., Kordula, T., Maceyka, M., and Spiegel, S.: ORMDL proteins regulate ceramide levels during sterile inflammation, J. Lipid Res., 57, 1412–1422, https://doi.org/10.1194/jlr.M065920, 2016.
Clop, A., Vidal, O., and Amills, M.: Copy number variation in the genomes of domestic animals, Anim. Genet., 43, 503–517, https://doi.org/10.1111/j.1365-2052.2012.02317.x, 2012.
Fadista, J., Thomsen, B., Holm, L. E., and Bendixen, C.: Copy number variation in the bovine genome, BMC Genomics, 11, 284, https://doi.org/10.1186/1471-2164-11-284, 2010.
Fontanesi, L., Martelli, P. L., Beretti, F., Riggio, V., Dall'Olio, S., Colombo, M., Casadio, R., Russo, V., and Portolano, B.: An initial comparative map of copy number variations in the goat (Capra hircus) genome, BMC Genomics, 11, 639, https://doi.org/10.1186/1471-2164-11-639, 2010.
Fowler, K. E., Pong-Wong, R., Bauer, J., Clemente, E. J., Reitter, C. P., Affara, N. A., Waite, S., Walling, G. A., and Griffin, D. K.: Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds, BMC Genomics, 14, 784, https://doi.org/10.1186/1471-2164-14-784, 2013.
Gu, W., Zhang, F., and Lupski, J. R.: Mechanisms for human genomic rearrangements, Pathogenetics, 1, 4, https://doi.org/10.1186/1755-8417-1-4, 2008.
Henrichsen, C. N., Chaignat, E., and Reymond, A.: Copy number variants, diseases and gene expression, Hum. Mol. Genet., 18, R1–R8, https://doi.org/10.1093/hmg/ddp011, 2009.
Hjelmqvist, L., Tuson, M., Marfany, G., Herrero, E., Balcells, S., and Gonzalez-Duarte, R.: ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins, Genome Biol., 3, research0027.1, https://doi.org/10.1186/gb-2002-3-6-research0027, 2002.
Hou, G. Y., Yuan, Z. R., Zhou, H. L., Zhang, L. P., Li, J. Y., Gao, X., Wang, D. J., Gao, H. J., and Xu, S. Z.: Association of thyroglobulin gene variants with carcass and meat quality traits in beef cattle, Mol. Biol. Rep., 38, 4705–4708, https://doi.org/10.1007/s11033-010-0605-1, 2011.
Kalman, B. and Vitale, E.: Structural chromosomal variations in neurological diseases, Neurologist, 15, 245–253, https://doi.org/10.1097/NRL.0b013e3181963cef, 2009.
Kim, Y. K., Moon, S., Hwang, M. Y., Kim, D. J., Oh, J. H., Kim, Y. J., Han, B. G., Lee, J. Y., and Kim, B. J.: Gene-based copy number variation study reveals a microdeletion at 12q24 that influences height in the Korean population, Genomics, 101, 134–138, https://doi.org/10.1016/j.ygeno.2012.11.002, 2013.
Lehnert, S. A., Reverter, A., Byrne, K. A., Wang, Y., Nattrass, G. S., Hudson, N. J., and Greenwood, P. L.: Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds, BMC Dev. Biol., 7, 95, https://doi.org/10.1186/1471-213x-7-95, 2007.
Lin, S., Lin, X., Zhang, Z., Jiang, M., Rao, Y., Nie, Q., and Zhang, X.: Copy Number Variation in SOX6 Contributes to Chicken Muscle Development, Genes (Basel), 9, 42, https://doi.org/10.3390/genes9010042, 2018.
Ma, Q., Liu, X., Pan, J., Ma, L., Ma, Y., He, X., Zhao, Q., Pu, Y., Li, Y., and Jiang, L.: Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array, Sci. Rep.-UK, 7, 912, https://doi.org/10.1038/s41598-017-00847-9, 2017.
McKnight, A. J., Currie, D., and Maxwell, A. P.: Unravelling the genetic basis of renal diseases; from single gene to multifactorial disorders, J. Pathol., 220, 198–216, https://doi.org/10.1002/path.2639, 2010.
Pinto, D., Darvishi, K., Shi, X., Rajan, D., Rigler, D., Fitzgerald, T., Lionel, A. C., Thiruvahindrapuram, B., Macdonald, J. R., Mills, R., Prasad, A., Noonan, K., Gribble, S., Prigmore, E., Donahoe, P. K., Smith, R. S., Park, J. H., Hurles, M. E., Carter, N. P., Lee, C., Scherer, S. W., and Feuk, L.: Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants, Nat. Biotechnol., 29, 512–520, https://doi.org/10.1038/nbt.1852, 2011.
Siow, D. L. and Wattenberg, B. W.: Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis, J. Biol. Chem., 287, 40198–40204, https://doi.org/10.1074/jbc.C112.404012, 2012.
Takumi, T. and Tamada, K.: CNV biology in neurodevelopmental disorders, Curr. Opin. Neurobiol., 48, 183–192, https://doi.org/10.1016/j.conb.2017.12.004, 2018.
Toncheva, A. A., Potaczek, D. P., Schedel, M., Gersting, S. W., Michel, S., Krajnov, N., Gaertner, V. D., Klingbeil, J. M., Illig, T., Franke, A., Winkler, C., Hohlfeld, J. M., Vogelberg, C., von Berg, A., Bufe, A., Heinzmann, A., Laub, O., Rietschel, E., Simma, B., Genuneit, J., Muntau, A. C., and Kabesch, M.: Childhood asthma is associated with mutations and gene expression differences of ORMDL genes that can interact, Allergy, 70, 1288–1299, https://doi.org/10.1111/all.12652, 2015.
Wang, S., Robinet, P., Smith, J. D., and Gulshan, K.: ORMDL orosomucoid-like proteins are degraded by free-cholesterol-loading-induced autophagy, P. Natl. Acad. Sci. USA, 112, 3728–3733, https://doi.org/10.1073/pnas.1422455112, 2015.
Wang, X., Nahashon, S., Feaster, T. K., Bohannon-Stewart, A., and Adefope, N.: An initial map of chromosomal segmental copy number variations in the chicken, BMC Genomics, 11, 351, https://doi.org/10.1186/1471-2164-11-351, 2010.
Wang, Y., Gu, X., Feng, C., Song, C., Hu, X., and Li, N.: A genome-wide survey of copy number variation regions in various chicken breeds by array comparative genomic hybridization method, Anim. Genet., 43, 282–289, https://doi.org/10.1111/j.1365-2052.2011.02308.x, 2012.
Xu, Y., Shi, T., Cai, H., Zhou, Y., Lan, X., Zhang, C., Lei, C., Qi, X., and Chen, H.: Associations of MYH3 gene copy number variations with transcriptional expression and growth traits in Chinese cattle, Gene, 535, 106–111, https://doi.org/10.1016/j.gene.2013.11.057, 2014.
Xu, Y., Zhang, L., Shi, T., Zhou, Y., Cai, H., Lan, X., Zhang, C., Lei, C., and Chen, H.: Copy number variations of MICAL-L2 shaping gene expression contribute to different phenotypes of cattle, Mamm. Genome, 24, 508–516, https://doi.org/10.1007/s00335-013-9483-x, 2013.
Yang, F., Cao, P., and Zhou, G.: Association of common copy number variations with diseases, Chinese Journal of Medical Genetics, 33, 388–391, https://doi.org/10.3760/cma.j.issn.1003-9406.2016.03.025, 2016.
Yang, L., Wang, Y. Z., Zhu, H. H., Chang, Y., Li, L. D., Chen, W. M., Long, L. Y., Zhang, Y. H., Liu, Y. R., Lu, J., and Qin, Y. Z.: PRAME Gene Copy Number Variation Is Related to Its Expression in Multiple Myeloma, DNA Cell Biol., 36, 1099–1107, https://doi.org/10.1089/dna.2017.3951, 2017.
Yang, L., Xu, L., Zhou, Y., Liu, M., Wang, L., Kijas, J. W., Zhang, H., Li, L., and Liu, G. E.: Diversity of copy number variation in a worldwide population of sheep, Genomics, 110, 143–148, https://doi.org/10.1016/j.ygeno.2017.09.005, 2018a.
Yang, L., Xu, L., Zhou, Y., Liu, M., Wang, L., Kijas, J. W., Zhang, H., Li, L., and Liu, G. E.: Diversity of copy number variation in a worldwide population of sheep, Genomics, 110, 143–148, https://doi.org/10.1016/j.ygeno.2017.09.005, 2018b.
Yang, M., Lv, J., Zhang, L., Li, M., Zhou, Y., Lan, X., Lei, C., and Chen, H.: Association study and expression analysis of CYP4A11 gene copy number variation in Chinese cattle, Sci. Rep.-UK, 7, 46599, https://doi.org/10.1038/srep46599, 2017.
Zhang, G. M., Zheng, L., He, H., Song, C. C., Zhang, Z. J., Cao, X. K., Lei, C. Z., Lan, X. Y., Qi, X. L., Chen, H., and Huang, Y. Z.: Associations of GBP2 gene copy number variations with growth traits and transcriptional expression in Chinese cattle, Gene, 647, 101–106, https://doi.org/10.1016/j.gene.2018.01.004, 2018.
Zhao, Y. X. and Li, M. H.: Research advances on the origin, evolution and genetic diversity of Chinese native sheep breeds, Hereditas (Beijing), 39, 958–973, https://doi.org/10.16288/j.yczz.17-102, 2017.
Zheng, L., Xu, J. W., Li, J. C., Wang, D. H., An, Q. M., Xu, L. N., Ma, Y. L., Wang, J., Peng, S. J., Lei, C. Z., Lan, X. Y., Chen, H., Huo, L. J., and Huang, Y. Z.: Distribution and association study in copy number variation of KCNJ12 gene across four Chinese cattle populations, Gene, 689, 90–96, https://doi.org/10.1016/j.gene.2018.12.019, 2019.