the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
A 20-bp insertion/deletion (indel) polymorphism within the CDC25A gene and its associations with growth traits in goat
Wenbo Cui
Nuan Liu
Xuelian Zhang
Yanghai Zhang
Lei Qu
Hailong Yan
Xianyong Lan
Wuzi Dong
Chuanying Pan
Cell division cycle 25A (CDC25A), a member of the CDC25 family of phosphatases, is required for progression from G1 to the S phase of the cell cycle. CDC25A provides an essential function during early embryonic development in mice, suggesting that it plays an important role in growth and development. In this study, we used mathematical expectation (ME) methods to identify a 20-bp insertion/deletion (indel) polymorphism of CDC25A gene in Shaanbei White Cashmere (SBWC) goats. We also investigated the association between this 20-bp indel and growth-related traits in SBWC goats. Association results showed that the indel was related to growth traits (height at hip cross, cannon circumference, and cannon circumference index) in SBWC goats. The height at hip cross of individuals with insertion/insertion (II) genotype was higher than those with insertion/deletion (ID) genotype (P=0.02); on the contrary, the cannon circumference and cannon circumference index of individuals with ID genotype were superior when compared with those with II genotype (P=0.017 and P=0.009). These findings suggest that the 20-bp indel in the CDC25A gene significantly affects growth-related traits, and could be utilized as a candidate marker for marker-assisted selection (MAS) in the cashmere goat industry.
- Article
(960 KB) - Full-text XML
- BibTeX
- EndNote
Goat is one of the most important livestock species and the oldest economic domesticated species, being used for meat, milk, and cashmere all over the world. Also, goat is characterized by its high reproduction rate, high quality of meat, strong compliance, and easy management, and is widely cultivated on a global scale, especially in China, India, and Pakistan (Liu and Zhou., 2015). With the improvement of society, goat meat is gradually consumed by the masses because of its high protein, low fat, and low cholesterol (Mao et al., 2012). Shaanbei White Cashmere (SBWC) goat is a cashmere and meat type of goat. Because of the high quality of meat, SBWC goats have been the main economic variety in Yulin city, Shaanxi province. In order to improve the comprehensive economic benefits of SBWC goats, it is necessary to study the growth and development traits so that we are able to lay a foundation for vigorously developing the meat performance of cashmere goats.
Goat production relies mainly on grazing on communal lands that hardly provide the minimum nutrient requirements due to overstocking and degradation in Asia, Africa, and Latin America. Appropriate breeding strategies should be designed to promote conservation and improvement of goat unique attributes (Escareño et al., 2013). The molecular marker-assisted selection (MAS), which is commonly used in molecular breeding, has become an important part of modern breeding technology systems. The principle of the technology is to use the molecular markers or functional markers closely linked to the target gene to accurately identify the genotypes of different individuals in the hybrid progeny, and to carry out the breeding technique based on the assisted selection (Bai et al., 2018). At present, MAS based on relevant genetic variants is used extensively to improve traits with low heritability, such as those associated with growth and reproduction (Silpa et al., 2018; Zhang et al., 2018). Single-nucleotide polymorphism (SNP), insertion/deletion (indel) and structural variation (SV) are the major genetic variations (Mullaney et al., 2010) and the main kinds of MAS. It was reported that a multiallelic indel in the promoter region of the Cyclin-dependent Kinase Inhibitor 3 gene was significantly associated with body weight and carcass traits in chickens (Li et al., 2018). A 10-bp indel polymorphism in the bovine PAX7 promoter altered the binding of the transcriptional factor ZNF219 and modulated promoter activity and gene expression in Chinese cattle (Xu et al., 2018). Also, some literature illustrated that both SNP and indel variations were closely related to certain traits, including the growth traits of cashmere goat (Zhang et al., 2015; Wang et al., 2019a). However, major genes affecting the growth traits of goats have not been found, and further research is needed.
Currently, whole-genome sequencing and genome-wide association studies (GWASs) are used to explore genetic variants strongly associated with production traits (Wang et al., 2016; Rahmatalla et al., 2018; Yang et al., 2018). In 2016, a study using whole-genome analysis identified several genes, which may have contributed to the phenotypes in body size in goat populations from eight domesticated goat breeds, as potentially critical for fecundity, including cell division cycle 25A (CDC25A) (Wang et al., 2016). CDC25A is a member of the CDC25 family of phosphatases and also a dual-specificity protein phosphatase. Several literature studies have shown that CDC25A is one of the most crucial cell cycle regulators which is required for activation of the apoptotic cell cycle pathway (Shen and Huang, 2012; Biswas et al., 2017). In terms of embryos, promoting CDC25A expression can regulate cell proliferation and axis extension during gastrulation in zebrafish. CDC25A ensured the health and genomic stability of the developing embryo in mice (Lee et al., 2009; Liu et al., 2017). However, the relationship between CDC25A and growth traits has never been reported in goat. Therefore, it is essential to explore the association between the CDC25A gene and the growth traits of SBWC goats.
In this study, a 20-bp indel polymorphism of the CDC25A gene was found in SBWC goat by using mathematical expectation (ME) methods (Yang et al., 2016). Furthermore, this 20-bp indel polymorphism of the CDC25A gene was found to be associated with the growth traits of SBWC goats. Our findings provide a basis for further research on the underlying causal mutation and suggest hypotheses for further studies leading to the application of MAS for goat breeding.
All experimental procedures were approved by the Review Committee for the Use of Animal Subjects of Northwest A&F University, China. The animal experimentation, including sample collection, was performed in agreement with the guidelines of the ethics commission.
2.1 Animals and data collection
All experimental animals were raised at the SBWC breeding farmland and managed under the same conditions. A total of 729 ear tissue samples from female SBWC goats were collected randomly from the SBWC breeding farm in Yulin, Shaanxi province (Wang et al., 2018a, 2019b). Growth traits for all selected unrelated individuals were measured, including body height (BH), body length (BL), heart girth (HG), chest depth (ChD), chest width (ChW), height at hip cross (HHC), and cannon circumference (CC); consequently, body length index (BLI), heart girth index (HGI), chest width index (ChWI), cannon circumference index (CCI), and body trunk index (BTI) were also calculated on the basis of our reported description (Jia et al., 2015; Yang et al., 2017). All tissues were stored at −80 ∘C until used for analysis and DNA experimentation.
2.2 DNA extraction and genomic DNA pools construction
The DNA was extracted using the high-salt extraction and phenol chloroform methods (Lan et al., 2007) and then diluted to a standard concentration (10 ng µL−1) and stored at −20 ∘C for the detection of genetic variation. The Nanodrop 1000 instrument was used to assess DNA purity (A260∕280 ratio) and quality.
A total of 50 DNA samples were randomly selected from each breed to construct genomic DNA pools. Genomic DNA samples were diluted to a standard 10 ng µL−1 concentration and individual aliquots of DNA samples were transferred to a single tube to ensure that a constant amount of each DNA sample was transferred to the pool (Yang et al., 2016; Li et al., 2017). The pool was then mixed gently and uniformly. The genomic DNA pools were used as a template for polymerase chain reaction (PCR) amplification and then PCR products were sequenced (Sham et al., 2002).
2.3 Primer design and PCR amplification
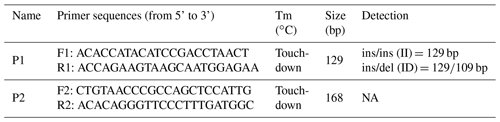
Based on the goat (Capra hircus) gene sequence (GenBank accession no. NC_030829.1) and the Ensembl database (http://www.ensembl.org/index.html?redirect=no, last access: 1 September 2018), two potential indel sites were found in CDC25A goat gene and two pairs of primers (P1, P2; Table 1) were designed to amplify genomic DNA pools to explore genetic variation in the CDC25A goat gene by the NCBI website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi, last access: 1 September 2018). The touch-down (TD) PCR reaction procedure was as follows: initial denaturation for 5 min at 95 ∘C followed by 18 cycles of denaturation for 30 s at 94 ∘C; annealing for 30 s at 68 ∘C (with a decrease of 1 ∘C per cycle); extension for 30 s at 72 ∘C; another 23 cycles of 30 s at 94 ∘C, 30 s at 50 ∘C, and 2 min at 72 ∘C; and a final extension for 10 min at 72 ∘C with subsequent cooling to 4 ∘C (Yang et al., 2016; Kang et al., 2019). The PCR was performed in a 25 µL reaction volume containing 12.5 µL 2× Taq Master mix, 0.5 µL of each primer, 2 µL 10 ng µL−1 genomic DNA and 9.5 µL ddH2O (double-distilled H2O).
2.4 Indel identification and sequencing
According to previous reports, based on the low frequencies of the 20-bp indel within the CDC25A gene and the sample sizes, we designed the most efficient pooling strategy to detect all the individuals by using the ME method (Yang et al., 2016). The PCR products specificity was confirmed by sequencing (Nakamura et al., 2007). The 20-bp indel of CDC25A was detected in SBWC breeds by electrophoresis using 3.5 % agarose gel stained with ethidium bromide.
2.5 Statistical analysis
Genotypic and allelic frequencies were calculated directly. The χ2 test was carried out to test whether the polymorphism is in Hardy–Weinberg equilibrium (HWE). Polymorphism information content (PIC) was calculated by Nei's method implemented in the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx, last access: 1 December 2018) (Cui et al., 2018). Difference distributions of genotypic and allelic frequencies were analyzed using the χ2 test or Fisher exact tests (when the minimum theoretical frequency was less than 5) in SPSS (version 19.0). For growth traits, analysis of variance was applied to the general linear model and the reduced linear model was as follows: , where Yijk is the observation of the growth trait (body height, etc.) evaluated on the ith level of the fixed factor age (αi) and the jth level of the fixed factor genotype (βj), μ is the overall mean for each trait, and εijk is the random error for the i, j, and kth individual (Wang et al., 2017). Further analysis was performed with SPSS 19.0 software using t test and Mann–Whitney U test; the data were rejected when n<5; P<0.05 was considered statistically significant.
3.1 PCR amplification and sequencing of the 20-bp indel variants of the CDC25A goat gene
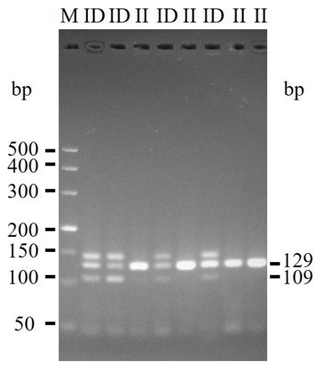
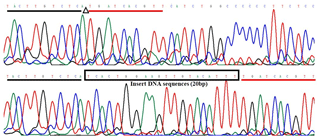
The 20-bp indel of the CDC25A goat gene was identified by P1 (Table 1) and genotyped by the 3.5 % agarose gel electrophoresis. However, the indel was not identified by P2 (Table 1). For the 20-bp indel locus, only genotypes insertion/insertion (II) and insertion/deletion (ID) were detected: one band (129 bp) for genotype II, and three bands (129/109 bp) and another homoduplex for genotype ID (Fig. 1). Herein, a 20-bp indel in the ninth intron of CDC25A goat gene was confirmed (NC_030829.1:g.51746323-51746342delTCACTGGAAGTTGTACATTT). The result of contrasting and analyzing the sequence by software (Bioedit, UK) showed that the indel sequence was “TCACTGGAAGTTGTACATTT” (Fig. 2).
3.2 Individuals genotyping by ME method
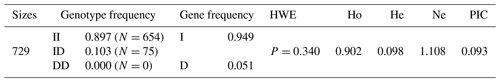
The low frequency of the 20-bp indel was confirmed in CDC25A gene in SBWC goat. Using the ME method, individuals were assigned by order in groups (the least allowed number in a single group) to mixed groups. Dependent on whether there was one single band (129 bp or 109 bp) in the mixed groups of SBWC goat, we needed to detect the genotype. Simultaneously, the number of PCR reactions was decreased. Results showed that the allelic frequencies of I and D were 0.949 and 0.051, respectively. Also, this indel locus was not in HWE and low polymorphic with a polymorphism information content (PIC) (Table 2).
3.3 Association of the indel locus and growth-related traits of SBWC goat
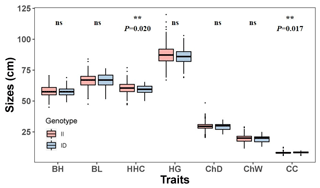
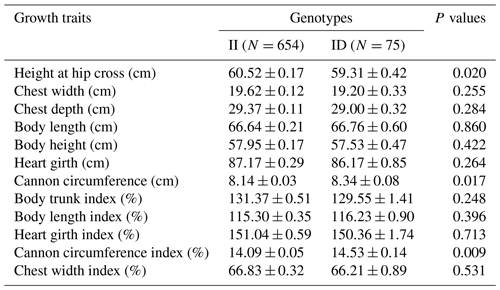
The association between the 20-bp indel of CDC25A gene and the growth traits were investigated in the SBWC goat breeds (Tables 3, 4; Fig. 3). Significant relationships were observed between this indel locus and cannon circumference (P=0.017), and cannon circumference index (P=0.009) in SBWC goat. This indel locus also appeared to have an approximate effect on other traits such as height at hip cross (P=0.020). The height at hip cross of individuals with II genotype was higher than those with ID genotype (P=0.02); on the contrary, the cannon circumference and cannon circumference index of individuals with ID genotype were superior when compared with those with II genotype (P=0.017 and P=0.009). Besides, analysis results showed that there was no significant correlation between the other growth traits and this indel locus in SBWC goat.
Table 3Relationship between the 20-bp indel locus of CDC25A gene and growth-related traits in SBWC goat (least square mean, LSMa ± SE).
CDC25A is specifically degraded in response to DNA damage, which prevents cells with chromosomal abnormalities from progressing through cell division. As mentioned in the literature, CDC25A dephosphorylates cyclin-dependent kinase and regulates the cell cycle in cell proliferation (Liang et al., 2016). In zebrafish, upon misexpression of CDC25A, several essential T-box transcription factors are abnormally expressed, which specifically prevents the normal onset of myoD transcription, leading to aberrant muscle formation (Bouldin et al., 2014). CDC25A can promote cell proliferation and osteoblast differentiation and ensure the health and genomic stability of the developing embryo (Verduzco et al., 2012; Qiu and Kassem, 2014). These scientific research efforts confirmed the importance of CDC25A in terms of growth and development. MAS is the most common method in molecular breeding. Especially indel has been widely reported in animal breeding for potential MAS (Ren et al., 2017; Cui et al., 2018; Zhao et al., 2018; Kang et al., 2019; Yang et al., 2019). Therefore, we aimed to determine the relationship between the indel polymorphism within the CDC25A and growth traits in goat.
Interestingly, we not only detected the II and ID genotypes, but also found a nontarget band in the CDC25A gene (Fig. 1). In fact, there were many similar studies about the nontarget fragment (Lu et al., 2014; Ren et al., 2017). Those nontarget bands were ultimately identified as heteroduplexes. In 1989, Nagamine et al. demonstrated that the generation of heteroduplexes theoretically occurred in any PCR reaction in which the genomic DNA carried an indel mutation. And if the heteroduplexes would be detected in the indel genes, it just existed in heterozygotes individuals (Nagamine et al., 1989). Heterozygotes in the present study showed the presence of heteroduplexes, which is consistent with the previous studies.
We firstly used an ME strategy to detect the allele frequency in all individuals (Yang et al., 2016). The present study confirmed the low frequency of this indel by randomly detecting 50 individuals one by one, and we established the most efficient pooling strategy based on the ME method. Finally, a total of 366 reaction times were performed in SBWC goat. Obviously, comparing with the one-by-one detecting method, the real times of ME-method PCR were considerably decreased. Doubtless, the successful application of ME method in our study was also consistent with previous studies (Yang et al., 2016; Li et al., 2017). Furthermore, our results found that the 20-bp indel of CDC25A goat gene was not in HWE in SBWC goat(P<0.05).
Furthermore, this study is the first report of the association between the 20-bp indel in the ninth intron of the CDC25A gene and the growth traits in SBWC goat. We found that individuals with genotype II were superior in higher height at hip cross, while they were inferior in cannon circumference and the cannon circumference index (Tables 3, 4; Fig. 3). Why? The feeding method of SBWC goats has changed from the traditional production mode of grazing to the production mode based on house feeding. Due to changes in feeding methods, some body size traits of goats have improved significantly, one of them is cannon circumference (Tan et al., 2012; Zhang et al., 2012). Therefore, we considered that goats needed bigger cannon circumference to support weight due to the needs of goat meat. As mentioned in the literature, weight gain rate of Shaanbei White Cashmere goat is relatively fast at the ages of 1 month and 4–5 months, and growth rates of body measurement indexes were relatively fast at the ages of 4–5 months and 7–9 months (Huang et al., 2017). We speculated that this discrepancy could be attributed to the lack of nutrition during development. Moreover, we did not find any individual with genotype DD in the study. We speculated that the mutation frequency of genotype DD was too low to detect.
Table 4Hypothesis test summary for relationship between the genotypes from the 20-bp indel locus of CDC25A gene and growth traits in SBWC goat (Mann–Whitney U test).
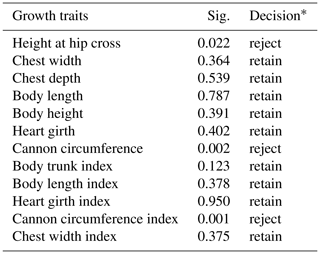
Note: Decision*: reject means that we reject the null hypothesis and retain means that we retain the null hypothesis. The null hypothesis is that the distribution of each growth trait is the same across categories of genotype. Asymptotic significance values (Sig.) are displayed. The significance level is 0.05.
In addition, although we found that this 20-bp indel was in the ninth intron of the CDC25A gene, the intron might also affect the phenotypic traits, which is consistent with previous reports. For example, a novel 43-bp indel polymorphism in intron 1 of the heparan sulfate 6-O-sulfotransferase 3 (HS6ST3) gene is significantly associated with growth and carcass traits in chickens (Wang et al., 2018b). It is also reported that a nucleotide substitution in intron 3 of IGF2 causes a major quantitative trait loci (QTLs) effect on muscle growth in pig (Van Laere et al., 2003). Therefore, we speculated that this 20-bp indel might also affect growth traits in SBWC goats.
In summary, an economic ME method was presented to quickly and accurately detect a low frequency of mutation, such as the 20-bp indel in the ninth intron of the CDC25A gene, which can save time and reduce expenses. Besides, the detected 20-bp indel significantly affects growth traits, which might be a potential useful DNA marker for MAS in SBWC goat.
The original data of the paper are available from the corresponding author upon request.
WC edited and revised the paper. NL and XZ implemented and collected the data. YZ analyzed the results and revised the paper. LQ and HY provided samples of SWBC goats. WD revised the paper. XL and CP designed the experiment. All authors reviewed and approved the final paper.
The authors declare that they have no conflict of interest.
This study was supported by the National Natural Science Foundation in China (nos. 31702115; 31760650) and Key Projects of Shaanxi province (2014KTDZ02-01). We are very grateful to Xiaoyue Song, Jinwang Liu, and Haijing Zhu (in Shaanxi Provincial Engineering and Technology Research Center of Cashmere Goats, Yulin University, and Life Science Research Center, Yulin University) for the sample collection of SBWC goats. We sincerely thank Wenxian Zeng, at the College of Animal Science and Technology, Northwest A&F University, for providing experimental instruments and equipment. We greatly thank the staff of SBWC breeding farm, Shaanxi province, P. R. China, for collecting samples.
This research has been supported by the National Natural Science Foundation in China (grant no. 31702115) and the National Natural Science Foundation in China (grant no. 31760650).
This paper was edited by Steffen Maak and reviewed by Memis Ozdemir, Sihem Amiri, and one anonymous referee.
Bai, S., Yu, H., Wang, B., and Li, J.: Retrospective and perspective of rice breeding in China, J. Genet. Genomics, 45, 603–612, 2018.
Biswas, S. C., Sanphui, P., Chatterjee, N., Kemeny, S., Greene, L. A.: Cdc25A phosphatase: a key cell cycle protein that regulates neuron death in disease and development, Cell Death Dis., 8, e2692, https://doi.org/10.1038/cddis.2017.115, 2017.
Bouldin, C. M., Snelson, C. D., Farr III, G. H., and Kimelman, D.: Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body, Genes Dev., 28, 384–395, 2014.
Cui, Y., Yan, H., Wang, K., Xu, H., Zhang, X., Zhu, H., Liu, J., Qu, L., Lan, X., and Pan, C.: Insertion/deletion within the KDM6A gene is significantly associated with litter size in goat, Front Genet., 9, 91, https://doi.org/10.3389/fgene.2018.00091, 2018.
Escareño, L., Salinas-Gonzalez, H., Wurzinger, M., Iñiguez, L., Sölkner, J., and Meza-Herrera, C.: Dairy goat production systems: status quo, perspectives and challenges, Trop. Anim. Health Prod., 45, 17–34, 2013.
Huang, S., Shi, L., Bi, T. F., Zhu, H. J., Yu, H. H., and Qu, L.: Growth and development regulation of Shaanbei white cashmere goat, Chinese Agricultural Science Bulletin, 26, 107–111, 2017.
Jia, W. C., Wu, X. F., Li, X. C., Xia, T., Lei, C. Z., Chen, H., Pan, C. Y., and Lan, X. Y.: Novel genetic variants associated with mRNA expression of signal transducer and activator of transcription 3 (STAT3) gene significantly affected goat growth traits, Small Ruminant Res., 129, 25–36, 2015.
Kang, Z. H., Jiang, E. H., Wang, K., Pan, C. Y., Chen, H., Yan, H. L., Zhu, H. J., Liu, J. W., Qu, L., and Lan, X. Y.: Goat membrane associated ring-CH-type finger 1 (MARCH1) mRNA expression and association with litter size, Theriogenology, 128, 8–16, 2019.
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., Lei, C. Z., Zhang, A. L., and Zhang, L.: An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits, Small Ruminant Res., 73, 8–12, 2007.
Lee, G., White, L. S., Hurov, K. E., Stappenbeck, T. S., and Piwnica-Worms, H.: Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion, P. Natl. Acad. Sci. USA, 106, 4701–4706, 2009.
Li, J., Zhu, X., Ma, L., Xu, H., Cao, X., Luo, R., Chen, H., Sun, X., Cai, Y., and Lan, X.: Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method, Prion, 11, 143–150, 2017.
Li, W., Liu, D., Tang, S., Li, D., Han, R., Tian, Y., Li, H., Li, G., Li, W., Liu, X., Kang, X., and Li, Z.: A multiallelic indel in the promoter region of the Cyclin-dependent kinase inhibitor 3 gene is significantly associated with body weight and carcass traits in chickens, Poult. Sci., 98, 556–565, 2018.
Liang, J., Cao, R., Zhang, Y., Xia, Y., Zheng, Y., Li, X., Wang, L., Yang, W., and Lu, Z.: PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis, Nat. Commun., 7, 12431, https://doi.org/10.1038/ncomms12431, 2016.
Liu, K. W. and Zhou, Y.: Discussion on rural goat breeding technology, Livestock and Poultry Industry, 8, 26–27, https://doi.org/10.19567/j.cnki.1008-0414, 2015.
Liu, Y., Sepich, D. S., and Solnica-Krezel, L.: Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation, PLoS Genet., 13, e1006564, https://doi.org/10.1371/journal.pgen.1006564, 2017.
Lu, P., Feng, X., Zhou, J. S., Liu, X. B., Chen, Q. X., and Li, M.: Identification and analysis of a 5-bp indel of a porcine BMP7 gene promoter, Genet. Mol. Res., 13, 4326–4335, 2014.
Mao, J. W., Xu, H. Z., and Zhao, Y. J.: Development status of high-grade mutton and main points of production technology, Heilongjiang Animal Science and Veterinary Medicine, 10, 64–65, https://doi.org/10.13881/j.cnki.hljxmsy.2012.10.014, 2012.
Mullaney, J. M., Mills, R. E., Pittard, W. S., and Devine, S. E.: Small insertions and deletions (INDELs) in human genomes, Hum. Mol. Genet., 19, 131–136, 2010.
Nagamine, C. M., Chan, K., and Lau, Y. F.: A PCR antifact generation of heteroduplexes, Am. J. Hum. Genet., 45, 337–339, 1989.
Nakamura, I., Xue, G., Sakudo, A., Saeki, K., Matsumoto, Y., Ikuta, K., and Onodera, T.: Novel single nucleotide polymorphisms in the specific protein 1 binding site of the bovine PRNP promoter in Japanese Black cattle: impairment of its promoter activity, Intervirology, 50, 190–196, 2007.
Qiu, W. and Kassem, M.: miR-141-3p inhibits human stromal (mesenchymal) stem cell proliferation and differentiation, Biochim. Biophys. Acta., 1843, 2114–2121, 2014.
Ren, F., Yu, S., Chen, R., Lv, X. Y., and Pan, C. Y.: Identification of a novel 12-bp insertion/deletion (indel) of iPS-related Oct4 gene and its association with reproductive traits in male piglets, Anim. Reprod. Sci., 178, 59–64, 2017.
Rahmatalla, S. A., Arends, D., Reissmann, M., Wimmers, K., Reyer, H., and Brockmann, G. A.: Genome-wide association study of body morphological traits in Sudanese goats, Anim. Genet., 49, 478–482, 2018.
Sham, P., Bader, J. S., Craig, I., O'Donovan, M., and Owen, M.: DNA Pooling: a tool for large-scale association studies, Nat. Rev. Genet., 3, 862–871, 2002.
Shen, T. and Huang, S.: The role of Cdc25A in the regulation of cell proliferation and apoptosis, Anticancer Agents Med. Chem., 12, 631–639, 2012.
Silpa, M. V., Naicy, T., Aravindakshan, T. V., Radhika, G., Boswell, A., and Mini, M.: Sirtuin3 (SIRT3) gene molecular characterization and SNP detection in prolific and low prolific goat breeds, Theriogenology, 122, 47–52, 2018.
Tan, L., Gao, W. R., Wei, Q. Y., Tian, X. E., Wang, Y. J., and Chen, Y. L.: Study on body index changes with age for Shaanbei White Cashmere goat, Acta Ecologiae Animalis Domastici, 33, 35–38, 2012.
Van Laere, A. S., Nguyen, M., Braunschweig, M., Nezer, C., Collette, C., Moreau, L., Archibald, A. L., Haley, C. S., Buys, N., Tally, M., Andersson, G., Georges, M., and Andersson, L.: A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig, Nature, 425, 832–836, 2003.
Verduzco, D., Dovey, J. S., Shukla, A. A., Kodym, E., Skaug, B. A., and Amatruda, J. F.: Multiple isoforms of CDC25 oppose ATM activity to maintain cell proliferation during vertebrate development, Mol. Cancer Res., 10, 1451–1461, 2012.
Wang, K., Yan, H., Xu, H., Yang, Q., Zhang, S., Pan, C., Chen, H., Zhu, H., Liu, J., Qu, L., and Lan, X.: A novel indel within goat casein alpha S1 gene is significantly associated with litter size, Gene, pii, 30635-8, https://doi.org/10.1016/j.gene.2018.05.119.S0378-1119, 2018a.
Wang, K., Cui, Y., Wang, Z., Yan, H., Meng, Z., Zhu, H., Qu, L., Lan, X., and Pan, C.: One 16 bp insertion/deletion (indel) within the KDM6A gene revealing strong associations with growth traits in goat, Gene, 686, 16–20, 2019a.
Wang, X., Liu, J., Zhou, G., Guo, J., Yan, H., Niu, Y., Li, Y., Yuan, C., Geng, R., Lan, X., An, X., Tian, X., Zhou, H., Song, J., Jiang, Y., and Chen, Y.: Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits, Sci. Rep., 12, 38932, https://doi.org/10.1038/srep38932, 2016.
Wang, X., Yang, Q., Wang, K., Zhang, S., Pan, C., Chen, H., Qu, L., Yan, H., and Lan, X.: A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats, Anim. Genet., 48, 735–736, 2017.
Wang, X., Li, Z., Guo, Y., Wang, Y., Sun, G., Jiang, R., Kang, X., and Han, R.: Identification of a novel 43-bp insertion in the heparan sulfate 6-O-sulfotransferase 3 (HS6ST3) gene and its associations with growth and carcass traits in chickens, Anim. Biotechnol., 8, 1724, 2018b.
Wang, X., Yang, Q., Wang, K., Yan, H., Pan, C., Chen, H., Liu, J., Zhu, H., Qu, L., and Lan, X.: Two strongly linked single nucleotide polymorphisms (Q320P and V397I) in GDF9 gene are associated with litter size in cashmere goats, Theriogenology, 125, 115–121, 2019b.
Xu, Y., Shi, T., Zhou, Y., Liu, M., Klaus, S., Lan, X., Lei, C., and Chen, H.: A novel PAX7 10-bp indel variant modulates promoter activity, gene expression and contributes to different phenotypes of Chinese cattle, Sci. Rep., 8, 1724, 2018.
Yang, Q., Zhang, S. H., Liu, L. L., Cao, X. K., Lei, C. Z., Qi, X. L., Lin, F. P., Qu, W. D., Qi, X. S., Liu, J. M., Wang, R. M., Chen, H., and Lan, X. Y.: Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14-bp insertion/deletion (indel) within the bovine PRNP gene, Prion, 10, 409–419, 2016.
Yang, Q., Yan, H., Li, J., Xu, H., Wang, K., Zhu, H., Chen, H., Qu, L., and Lan, X.: A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size, Anim. Genet., 48, 499–500, 2017.
Yang, Q., Wu, P., Wang, K., Chen, D., Zhou, J., Ma, J., Li, M., Xiao, W., Jiang, A., Jiang, Y., Bai, L., Zhu, L., Li, X., and Tang, G.: SNPs associated with body weight and backfat thickness in two pig breeds identified by a genome-wide association study, Genomics, pii, S0888-7543(18)30329-X, https://doi.org/10.1016/j.ygeno.2018.11.002, 2018.
Yang, Q., Zhang, S., Li, J., Wang, X., Peng, K., Lan, X., and Pan, C.: Development of a touch-down multiplex PCR method for simultaneously rapidly detecting three novel insertion/deletions (indels) within one gene: an example for goat GHR gene, Anim. Biotechnol., 1, 1–6, https://doi.org/10.1080/10495398.2018.1517770, 2018.
Zhang, X., Wu, X., Jia, W., Pan, C., Li, X., Lei, C., Chen, H., and Lan, X.: Novel Nucleotide Variations, Haplotypes Structure and Associations with Growth Related Traits of Goat AT Motif-Binding Factor (ATBF1) Gene, Asian Austral. J. Anim., 28, 1394–1406, 2015.
Zhang, X., Yan, H., Wang, K., Zhou, T., Chen, M., Zhu, H., Pan, C., and Zhang, E.: Goat CTNNB1: mRNA expression profile of alternative splicing in testis and association analysis with litter size, Gene, 679, 297–304, 2018.
Zhang, Z. W., Hua, W. L., Ye, Y., Liu, Z. F., Shi, H. L., Ren, W. L., and Yan, H.: Regression analysis of body weight and body size of Zhongwei wethers under house feeding conditions, China Herbivore Science, 1, 417–418, 2012.
Zhao, H., He, S., Wang, S., Zhu, Y., Xu, H., Luo, R., Lan, X., Cai, Y., and Sun, X.: Two new insertion/deletion variants of the PITX2 gene and their Effects on growth traits in Sheep, Anim. Biotechnology, 29, 276–282, 2018.