the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Associations between genetic variants of the POU1F1 gene and production traits in Saanen goats
Güldehen Bilgen
This study was conducted to determine the polymorphisms of the POU1F1 gene and their relationships with milk yield and components, litter size, birth weight, and weaning weight in goats. For this purpose, a total of 108 Saanen goats from two different farms (Bornova and Manisa) were used as animal materials. Polymorphisms at the exon 6 and the 3′ flanking region of the POU1F1 gene were determined by using PCR-RFLP with PstI and AluI restriction enzymes and DNA sequencing analyses. Two alleles and three genotypes were identified by AluI or PstI digestions of the POU1F1 gene. The genotypes frequencies of TT, TC, and CC were 64.8 %, 31.5 % and 3.7 % for the PstI locus; 54.6 %, 31.5 % and 13.9 % for the AluI locus, respectively. T allele frequencies (0.56 and 0.88 for the AluI locus, 0.80 and 0.81 for the PstI locus, respectively) were predominant in both loci at the Bornova and Manisa farms. In terms of POU1F1-AluI and POU1F1-PstI loci, two populations were found to be in Hardy–Weinberg equilibrium. In the POU1F1-AluI locus, significant associations were found between genotypes and lactation milk yield and litter size. Similarly, a significant relationship between genotypes and birth weight in the POU1F1-PstI locus (p<0.05) was determined. The TC and CC genotypes were observed to be higher than the TT genotype for lactation milk yield and litter size at the POU1F1-AluI locus. Birth weight was found to be higher in animals that have the CC genotype at the POU1F1-PstI locus. In conclusion, the POU1F1 gene can be used as a molecular marker for economic features like reproduction, growth, milk content and yield in Saanen goats.
- Article
(1009 KB) - Full-text XML
-
Supplement
(246 KB) - BibTeX
- EndNote
Goat populations have increased in recent years in spite of changes in agriculture and technological progress in the world. Demand for goat milk and its products has increased because of an increase in health-conscious consumers. Goat milk provides great advantages to human nutrition such as high digestibility, being antiallergenic, and it has short- and medium-chain length fatty acids (Park et al., 2007; Getaneh et al., 2016). Therefore, goat breeding has become popular, and the goat population in Turkey has increased over the last 5 years (FAO, 2016).
Goat breeding is carried out in village flocks, plateaus or nomadic flocks in Turkey. However, in recent years in western Anatolia some entrepreneurs have invested money in intensive farming that provides goat milk to dairy farms. The Saanen breed and its hybrids are usually raised in such intensive farm enterprises (Kaymakçı and Dellal, 2006; Kaymakçı and Taşkın, 2006). It is known that the goat population is high in Çanakkale and Balikesir, especially in İzmir, in the western Anatolia region (FAO, 2016).
Genes related to economic traits and possible effects on production traits have been investigated using various DNA markers. One of these markers is single nucleotide polymorphisms (SNPs) that can occur in the forms of transitions or transversions. SNP markers are used for identifying genetic diversity, in quantitative trait locus (QTL) analysis and in genomic selection of livestock. Investigations have been performed to determine SNPs affecting characteristics such as resistance to mastitis and scrapie diseases, carcass and meat quality, milk yield, and milk fat and protein content because SNPs are a widespread polymorphism found in the genome, and are easy to identify (Li et al., 2011; Zhang et al., 2012; Corral et al., 2013; Wang et al., 2015; dos Santos et al., 2015; Paiva et al., 2016; Ekegbu et al., 2019).
POU1F1 (also called GHF-1 or PIT-1) is a member of the POU-domain family which is an important regulator for growth hormone (GH), prolactin (PRL), and thyroid-stimulating hormone β (TSHβ) (Tuggle and Trenkle, 1996; Cohen et al., 1997; Li et al., 2016). Many transcription factors are involved in pituitary organogenesis during development and maturation of the anterior pituitary gland. POU homeodomains such as PROP1 and POU1F1, and PITX homeodomains such as PITX2 and PITX1 have been associated with a decrease in GH and PRL expression, and with proliferation of somatotropic and lactotropic cell lines (Savage et al 2003; Huai et al., 2011; Selvaggi and Dario, 2011). The POU1F1 gene is located on 1q21–22 chromosome in goats, cattle and sheep and consists of 6 exons and 5 introns (Woollard et al., 2000).
The POU1F1 gene is an important candidate gene associated with growth, reproduction, milk yield, and milk components. This is because the POU1F1 transcription factor regulates the expression of genes GH, PRL, and TSHβ (Daga et al., 2013; Feng et al., 2012; Lan et al., 2009b, 2007a, b). Research has been carried out to investigate the association of the POU1F1 gene with milk yield, milk composition, and growth traits in goats and cattle (Lan et al., 2007a, b; Zhang et al., 2009; Zhou et al., 2016). Some of the polymorphisms in the POU1F1 gene were reported to be related to growth, weaning weight, litter size and meat quality traits in sheep (Mura et al., 2012; Özmen et al., 2013; Sadeghi et al., 2014; Jalil-Sarghale et al., 2014; Bai et al., 2016; AL-Khuzai and AL-Anbari 2018), milk production in cattle (Ahmadi et al., 2015), carcass weight in cattle (Seong et al., 2011), meat quality in rabbits (Wang et al., 2015), and milk production, growth traits and litter size in goats (Daga et al., 2013; Feng et al., 2012; Ma et al, 2017). Many studies have suggested that the POU1F1 gene may be a candidate gene to be used in marker-assisted selection programs (Feng et al., 2012; Ma et al, 2017; AL-Khuzai and AL-Anbari 2018).
This study aimed to investigate the polymorphisms of the POU1F1 gene and to evaluate their relationships with some characteristics of reproduction, growth, milk yield and milk components in Saanen goats that are reared in İzmir and Manisa Province.
2.1 Samples and DNA isolation
A total of 108 Saanen goats (60 goats reared in the Small Ruminant Animal Application and Research Unit, Ege University Faculty of Agriculture Department of Animal Science, Bornova; 48 goats reared in a private Saanen farm in Manisa Province) and their offspring were used as materials. A total of 162 offspring from 108 dams were used for litter size, and 88 offspring from 60 dams were used for birth weight and weaning weight.
The monthly milk yield of individuals was recorded twice a day during lactation in 2013–2014. Protein, fat and dry matter ratios were determined in milk samples with a Bentley 150 milk analyzer. 10 mL of blood sample from the vena jugularis of the 108 goats was collected in vacuum tubes containing K3 EDTA as anticoagulant. Genomic DNA was isolated using a commercial DNA isolation kit (K0721, GeneJET Whole Blood Genomic DNA Purification Mini Kit, Thermo Fisher Scientific) according to the manufacturer's protocol.
2.2 DNA amplification and genotyping
The exon 6 and 3′ flanking region of POU1F1 gene was amplified using F: 5′-CCATCATCTCCCTTCTT-3′ and R: 5′-AATGTACAATGTCCTTCTGAG-3)′ primers (Lan et al., 2007b). The 25 µL PCR volume contained 100 ng genomic DNA, 0.5 µM of each primers, 1× PCR Buffer, 200 µM dNTP, 2 mM MgCl2 and 1 U of Taq DNA polymerase (i-StarTaq™ DNA Polymerase, iNtRon Biotechnology). The cycling protocol was 5 min at 95 ∘C, 35 cycles of 94 ∘C for 30 s, 54 ∘C annealing for 30 s, 72 ∘C for 45 s with a final extension at 72 ∘C for 10 min.
PCR products of the POU1F1 gene were digested with 10 U of AluI and PstI restriction enzymes (FD0014 and FD0614, Thermo Fisher Scientific) at 37 ∘C for 3 h. PCR products and restriction fragments were electrophoresed on a 2.5 % agarose gel stained with SafeView™ Classic (Applied Biological Materials Inc., Canada).
POU1F1 gene fragments which gave different genotypes were also sequenced. The sequences of POU1F1 fragments were analyzed by using the MEGA6 software (Molecular Evolutionary Genetics Analysis, version 6.0; Tamura et al., 2013) for generating sequence alignments.
2.3 Statistical analysis
The genotypic and allelic frequencies of the POU1F1 gene and the Hardy–Weinberg equilibrium of the populations were calculated using the PopGene program (Yeh et al., 2000). The statistical software SPSS 18.0 was used to analyze the relationships between the genotypes and economic traits in goats.
Lactation milk yield was calculated according to the Trapeze II method, and the lactation period of 210 d was corrected to 280 d (ICAR, 2014). The total milk yield for each farm studied was statistically analyzed by the general linear model at significance level (α<0.05).
The adjusted linear model I with fixed effects was used to analyze the relationships between genotypes, milk yield, and components of 108 dairy goats. linear model I: , where Yijklm was the milk traits measured of each ijklmth animal, μ the overall mean, Bi the type of ith farm, Aj the jth lactation number, Gk the type of the kth genotype, Sl the type of the lth birth, and eijklm was the random error. The adjusted linear model II with fixed effects was used to analyze the relationships between genotype, birth weight, and weaning weight of 88 offspring. Linear model II: , where Yijkl was the weight traits measured on each of the ijklth animal, μ was the overall population mean, Si the type of birth of the ith offspring, Gj the type of the jth genotype, Ck the sex of the kth offspring (male, female), b the regression coefficient (Xijk; birth weight of dam, X; overall birth weight population mean of dam for birth weight, Xijk; weaning weight of offspring, X; overall weaning weight population mean of offspring for weaning weight), and eijkl was the random error. Effects associated with the farm and season of birth were not into incorporated into the linear model because the preliminary statistical analyses indicated that these effects did not have significant influences on the variability of traits in populations. The adjusted linear model III with fixed effects was used to analyze the relationships between genotype and litter size of 162 offspring. linear model III: , where Yijkl was the litter size trait measured for each ijklth animal, μ was the overall population mean, Ki the type of ith farm, Aj the jth lactation number, Gk the type of the kth genotype, and eijkl was the random error. Effects associated with the farm and season of birth were not incorporated into the linear model.
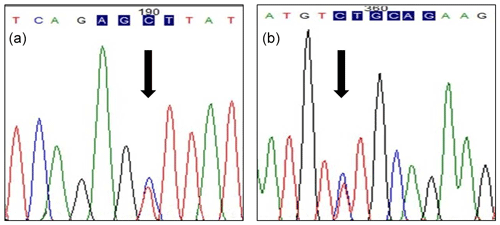
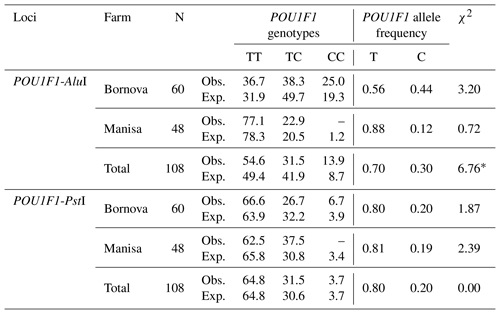
PCR–restriction fragment length polymorphism (PCR–RFLP) with AluI and PstI restriction enzymes and DNA sequencing were used to validate genetic polymorphism for exon 6 to the 3′ flanking region of the POU1F1 gene (450 bp) in two Saanen goat populations in Turkey. The transition from thymine to cytosine (DQ826413.1) (g.172T>C) in the sixth exon of the (210 bp long) POU1F1 gene was determined by the restriction enzyme AluI (Fig. 1a). The transition from cytosine to thymine (DQ826413.1) (g.110C>T) in the 3′ flanking region (total length 195 bp) was examined by the PstI restriction enzyme (Fig. 1b). Two alleles and three genotypes were identified with AluI (TT: 340, 110 bp; TC: 340, 216, 124, 110 bp; CC: 216, 124, 110 bp) and PstI (TT: 450 bp; TC: 450, 370, 80 bp; CC: 370, 80 bp) loci of the POU1F1 gene (Fig. 2a, b). The genotypes and allele frequencies of POU1F1 gene-AluI and POU1F1 gene-PstI analyses are listed in Table 1. POU1F1-T allele frequencies were 0.7 and 0.8 for POU1F1-AluI and POU1F1-PstI loci, respectively. The CC genotype was not found in Manisa farm for two loci. The POU1F1 gene sequenced that is identified in this study was deposited to the NCBI GenBank with the accession number MH892432.
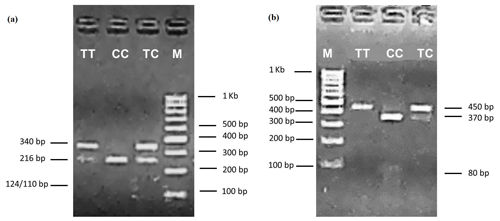
Figure 2Electrophoresis patterns of the POU1F1-AluI (a) and POU1F1-PstI (b) loci. TT, TC and CC are genotypes for the POU1F1-AluI and POU1F1-PstI loci, M; marker.
Table 1The genotypic and allelic frequencies of POU1F1-AluI and POU1F1-PstI analyses in Saanen dairy goats.
Similar to our results, the TT and TC genotypes were observed in the POU1F1-AluI locus in the Chinese indigenous breed, and the CC genotype was found to be very low in only two of the nine breeds (Lan et al., 2007b, 2009a). In POU1F1-PstI locus, genotype frequencies of TT and TC were observed as 91.7 % and 8.3 % respectively while CC genotype was not observed in native Chinese goats (Lan et al., 2009b). Also, as with Daga et al. (2013), TT and TC genotypes were observed in Italian goats and the CC genotype was not found. Similarly, it has been reported that the T allele is predominant in Chinese and Italian goats. The T allele was found to be monomorphic in Indian Barbari goats (Sharma et al., 2013).
Hershberg and Petrov (2008) reported that the tendency of codon usage may be different between codons of the same amino acid in different species, and that the frequency of the population may be lower in some populations as the codons reduce gene expression levels. The lack of each genotype POU1F1-AluI and POU1F1-PstI CC, and the presence of the small number of TC genotypes in the Manisa population, suggests that they may be related to the codon usage tendency, although the trends of codon usage in this study were not calculated.
In this study, all the genotype distributions of POU1F1-PstI are found in Hardy–Weinberg equilibrium (p>0.05) except for POU1F1-AluI (p<0.05). This can be because of random selection which is a result of artificial insemination. The reason for the absence of the CC genotype in the Manisa farm can be explained by the low number of samples. However, in some of the studies on the POU1F1-AluI loci in various goat breeds, it should be noted that the absence of the CC genotype is found at a very low frequency regardless of the number of samples (Daga et al., 2013; Lan et al., 2009a, b).
3.1 Associations between genetic variations of the POU1F1 gene and production traits
Individual milk yields of the Bornova and Manisa goats were recorded monthly, and protein, fat and dry matter ratios were determined in milk samples. Lactation milk yield was calculated as 731.94±14.17 kg, dry matter ratio 12.18±0.08, fat ratio 4.23±0.07 ,and protein ratio 3.31±0.02 in both farms (data not shown in table).
The relationships of the genotypes with milk yield and components for POU1F1-AluI and POU1F1-PstI loci are shown in Table 2. When two farms were evaluated together, TC and CC genotypes of POU1F1-AluI locus have higher milk yields (784.58 and 786.07 kg, respectively) than that of the TT genotype (p<0.05). According to Lan et al. (2007b), the sixth exon was associated with high milk yield and birth weight. Also, in previous studies it has been reported that g.102T>C and g.216T>C polymorphisms of POU1F1 gene are associated with the production traits such as milk yield and birth weight (Lan et al., 2007b, c). The relationships between genotypes, milk fat, protein ratio, and dry matter ratio in both farms were not found to be significant. The relationships between genotypes and milk fat ratio were not found to be significant, but the CC genotype in the POU1F1-AluI locus appeared to have the highest (4.44 %) milk fat ratio. The dry matter ratio of the TT genotype was found to be higher (12.87 %), which was statistically significant, than that of the TC genotype (12.24 %) in Manisa farm (p<0.05) for POU1F1-AluI locus (data not shown in Table 2). The relationships between genotypes and milk components were not found significant for POU1F1 ∕ PstI locus. Similar to our results, Zhou et al. (2016) reported that there was no significant relationship between POU1F1-PstI and milk performance in Guanzhong dairy goats. But they found that the TT genotype was higher than other genotypes for milk fat content and average milk fat.
Table 2Relationships of AluI and PstI polymorphisms of the POU1F1 gene with milk yield and milk components in Saanen dairy goats.
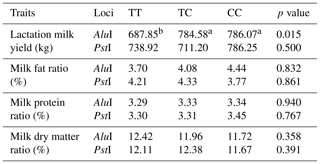
a,b Values with different superscripts within the same row differ significantly (p<0.05). n: 108 sample.
Relationships between the genotypes obtained from POU1F1-AluI, POU1F1-PstI and growth traits such as birth weight, weaning weight and litter size characteristics are shown in Table 3. The relationships between genotypes for POU1F1-AluI locus and birth weight and weaning weight were not found to be statistically significant at the Bornova farm. For POU1F1-AluI locus, the CC genotype was found higher in litter size than the TT and TC genotypes at the Bornova farm, whereas the CC genotype was not found at the Manisa Farm (p<0.05). When the two farms were evaluated together, the CC and TC genotypes were found higher in litter size than the TT genotype (p<0.01). In contrast with our results, Feng et al. (2012) reported that the litter size was higher in the case of the TT genotypes at C256T in exon 3 and G682T (A228S) in exon 6 of POU1F1 gene. According to Sun (2007), variants of the POU1F1 gene have significant influences on birth weight and weight at 1.5 years old on Liangshan sheep.
Table 3Relationships between the genotypes of POU1F1-AluI, POU1F1-PstI loci and growth traits.
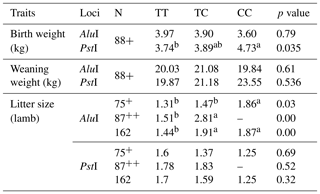
a,b Values with different superscripts within the same row differ significantly (p<0.05). + Bornova. Manisa sample number.
Differences between the CC, TC and TT genotypes (4.73, 3.89, and 3.74 kg, respectively) were found to be statistically significant (p<0.05) with the POU1F1-PstI locus for birth weight at the Bornova farm. For the POU1F1-AluI locus, birth weight was determined highest with the TT genotype, although it was not found to be statistically significant.
According to Ma et al. (2017), DQ826397.1:g.102T>G (SNP1), DQ826397.1:g.279T>C (SNP2) and NC_019460.2:g.1100T>A (SNP6) were associated with some growth traits such as hucklebone width, body weight, chest width, and chest circumference. The SNP1 locus at exon 6 had a significant association with hucklebone width (p<0.05) and the hucklebone width index (p<0.05) in Guanzhong dairy goats.
These results indicate that the POU1F1-AluI locus has significant effects on milk performance and litter size. Also the POU1F1-PstI locus has a significant effect on birth weight. Therefore, the SNPs of POU1F1 may be useful for potential marker-assisted selection in dairy goat breeding.
The POU1F1 gene is a transcription factor gene that plays a role in the regulation of expression of genes such as GH, PRL and TSHβ. It is also thought that POU1F1 may be a potential candidate gene for the marker-assisted selection of production traits such as milk yield, components, growth, and reproduction. Following the determination of polymorphisms, the determination of the mRNA expression level of the POU1F1 gene would be useful to reveal any indirect effects on the genes that could affect the quantitative traits.
The sequences of the samples studied are provided in the supplement.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-62-249-2019-supplement.
Both authors made substantial contributions to each step of the experimental procedure and manuscript preparation. GB supervised all stages of the experimental study. RI performed the sampling, the laboratory analysis and analyzed the molecular data. Both the authors wrote and prepared the manuscript.
The authors declare that they have no conflict of interest.
This paper is a summary of the PhD thesis of the first author, who was supported by TÜBİTAK-BİDEB 2211-C Priority Areas National Scholarship Program for PhD Students.
Animal welfare/ethical statement. The authors declare that this research has been carried out according to the laws and regulations of Turkey and fully compatible with the ethical guidelines of this journal. Furthermore, this study did not require any specific ethics committee approval considering the decision made by Animal Use and Ethical Committee of Ege University (2013-047)
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Ahmadi, M. M., Mirzaei, A., Sharifiyazdi, H., Hajibemani, A., and Ghasrodashti, R. A.: Pituitary-specific transcription factor 1 (Pit-1) polymorphism and its association on milk production and some reproductive performance in Holstein dairy cows, Rev. Med. Vet., 5–6, 127–131, 2015.
Al-Khuzai, H. M. and Al-Anbari, N. N.: Relationship of POU1F1 gene polymorphism with some of economical traits in Iraqi awassi ewes, Journal of Entomology and Zoology Studies, 6, 2082–2085, https://doi.org/10.22271/j.ento, 2018.
Bai, J. Y., Wang, X., Yang, Y. B., Zhang, X. H., Pang, Y. Z., and Li, H. W.: Study on the polymorphism of POU1F1 gene in sheep, Rev. Bras. Zootecn., 45, 604–607, https://doi.org/10.1590/S1806-92902016001000004, 2016.
Cohen, L. E., Wondisford, F. E., and Radovick, S.: Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin, Endocrin. Metab. Clin., 25, 523–540, https://doi.org/10.1016/S0889-8529(05)70339-X, 1997.
Corral, J. M., Padilla, J. A., Izquierdo, M., Martínez-Trancoón, M., Parejo, J. C., Salazar, J., and Hernández-García, F. I.: Detection and genetic characterization of ovine CSN1S2*B polymorphisms and their associations with milk production traits, Livest. Sci., 153, 10–19, https://doi.org/10.1016/j.livsci.2013.01.008, 2013.
Daga, C., Paludo, M., Luridiana, S., Consuelo Mura, M., Bodano, S., Pazzola, M., Dettori, M. L., Vacca, G. M., and Carcangiu, V.: Identification of novel SNPs in the Sarda breed goats POU1F1 gene and their association with milk productive performance, Mol. Biol. Rep., 40, 2829–2835, https://doi.org/10.1007/s11033-012-2298-0, 2013.
dos Santos, L. H., Lôbo, A. M. B. O., Facó, O., Gonçalves, H. C., and Lôbo, R. N. B.: Breeding programs for dairy goats generate profits in Brazil, Livest. Sci., 178, 27–34, https://doi.org/10.1016/j.livsci.2015.05.032, 2015.
Ekegbu, U. J., Burrows, L., Amirpour-Najafabadi, H., Zhou, H., and Hickford, J. G. H.: Gene polymorphisms in PROP1 associated with growth traits in sheep, Gene, 683, 41–46, https://doi.org/10.1016/j.gene.2018.10.024, 2019.
FAO: Food and Agriculture Organization of the United Nations, available at: http://www.fao.org/economic/ess/en/, last access: 10 June 2016.
Feng, T., Chu, M. X., Cao, G. L., Tang, Q. Q., Di, R., Fang, L., and Li, N.: Polymorphisms of caprine POU1F1 gene and their association with litter size in Jining Grey goats, Mol. Biol. Rep., 39, 4029–4038, https://doi.org/10.1007/s11033-011-1184-5, 2012.
Getaneh, G., Mebrat, A., Wubie, A., Kendie, H.: Review on Goat Milk Composition and Its Nutritive Value, Journal of Nutrition and Health Sciences, 3, 401, https://doi.org/10.15744/2393-9060.3.401, 2016.
Hershberg, R. and Petrov, D. A.: Selection on codon bias, Annu. Rev. Genet., 42, 287–99, https://doi.org/10.1146/annurev.genet.42.110807.091442, 2008.
Huai, Y. T., Lan, X. Y., Ma, L., Wang, J., Lei, C. Z., Chen, H., and Wang, J. Q.: Novel mutation in TGA stop-codon of bovine SIX6 gene, Mol. Biol., 45, 218–224, https://doi.org/10.1134/S0026893310061093, 2011.
ICAR: International Agreement of Recording Practices Book, General Assembly Berlin, Germany, 77 pp., 2014.
Jalil-Sarghale, A., Moradi Shahrbabak, M., Moradi Sharbabak, H., Sadeghi, M., and Mura, M. C.: Association of pituitary specific transcription factor-1 (POU1F1) gene polymorphism with growth and biometric traits and blood metabolites in Iranian Zel and Lori-Bakhtiari sheep, Mol. Biol. Rep., 41, 5787–5792, https://doi.org/10.1007/s11033-014-3451-8, 2014.
Kaymakçı, M. and Dellal, G.: Türkiye ve Dünya Keçi Yetiştiriciliği, Keçi Yetiştiriciliği, edited by: Kaymakçı, M., Meta Basım Matbaacılık Hizmetleri, İzmir İli Damızlık Koyun-Keçi Birliği Yayınları, İzmir, 2, 3–15, 2006.
Kaymakçı, M. and Taşkın, T.: Türkiye Süt Keçisi Geliştirme Yolları, Tayek/Tuyap Toplantısı25–27 Nisan, 2006 YılıHayvancılık Grubu Bilgi Alışveriş ToplantısıBildirileri, Yayın No 122, Menemen, İzmir, 2006.
Lan, X. Y., Pan, C. Y., Chen, H., Lei, C. Z., Hua, L. S., Yang, X. B., Qiu, G.Y., Zhang R. F., and Lun, Y. Z.: DdeI polymorphism in coding region of goat POU1F1 gene and its association with production traits, Asian-Austral. J. Anim., 20, 1342–1348, https://doi.org/10.5713/ajas.2007.1342, 2007a.
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., and Lei, C. Z., Zhang, A. L., and Zhang, L.: An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits, Small Ruminant Res., 73, 8–12, https://doi.org/10.1016/j.smallrumres.2006.10.009, 2007b.
Lan, X. Y., Pan, C. Y., Chen, H., and Lei, C. Z.: A DdeI PCR-RFLP detecting genetic variation of goat POU1F1 gene, Can. J. Anim. Sci., 87, 13–14, https://doi.org/10.4141/A06-068, 2007c.
Lan, X. Y., Li, M. J., Chen, H., Zhang, L. Z., Jing, Y. J., Wei, T. B., Ren, G., Wang, X., Fang, X. T., Zhang, C. L., and Lei, C. Z.: Analysis of caprine pituitary specific transcription factor-1 gene polymorphism in indigenous Chinese goats, Mol. Biol. Rep., 36, 705–709, https://doi.org/10.1007/s11033-008-9232-5, 2009a.
Lan, X. Y., Shu, J. H., Chen, H., Pan, C. Y., Lei, C. Z., Wang, X., Liu, S. Q., and Zhang, Y. B.: A PstI polymorphism at 3′UTR of goat POU1F1 gene and its effect on cashmere production, Mol. Biol. Rep., 36, 1371–1374, https://doi.org/10.1007/s11033-008-9322-4, 2009b.
Li, G., An, X. P., Fu, M. Z., Hou, J. X., Sun, R. P., Zhu, G. Q., Wang, J. G., and Cao, B. Y.: Polymorphism of PRLR and LHβ genes by SSCP marker and their association with litter size in Boer goats, Livest. Sci., 136, 281–286, https://doi.org/10.1016/j.livsci.2010.08.014, 2011.
Li, M. J., Zhang, C. M., Lan, X. Y., Fang, X. T., Lei, C. Z., and Chen, H.: Analysis of POU1F1 gene DdeI polymorphism in Chinese goats, Genet. Mol. Res., 15, 15017747, https://doi.org/10.4238/gmr.15017747, 2016.
Ma, L., Qin, Q., Yang, Q., Zhang, M., Zhao, H., Pan, C., Lei, C., Chen, H., and Lan, X.: Associations of six SNPs of POU1F1-PROP1-PITX1-SIX3 pathway genes with growth traits in two Chinese indigenous goat breeds, Ann. Anim. Sci., 17, 399–411, https://doi.org/10.1515/aoas-2016-0066, 2017.
Mura, C. M., Daga, C., Paludo, M., Luridiana, S., Pazzola, M., Bodano, S., Dettori, M. L., Vacca, G. M., and Carcangiu, V.: Analysis of polymorphism within POU1F1 gene in relation to milk production traits in dairy Sarda sheep breed, Mol. Biol. Rep., 39, 6975–6979, https://doi.org/10.1007/s11033-012-1525-z, 2012.
Özmen, Ö., Kul, S., and Ünal, E. O.: Polymorphism of sheep POU1F1 gene exon 6 and 3′UTR region and their association with milk production traits, Iran. J. Vet. Res., 15, 331–335, https://doi.org/10.22099/ijvr.2014.2587, 2013.
Paiva, S. R., McManus, C. M., and Blackburn, H.: Conservation of animal genetic resources – A new tact, Livest. Sci., 193, 32–38, https://doi.org/10.1016/j.livsci.2016.09.010, 2016.
Park, Y. W., Juarez, M., Ramos, M., and Haenlein, G. F. W.: Physico-chemical characteristics of goat and sheep milk, Small. Ruminant Res., 68, 88–113, https://doi.org/10.1016/j.smallrumres.2006.09.013, 2007.
Sadeghi, M., Jalil-Sarghale, A., and Moradi-Shahrbabak, M.: Associations of POU1F1 gene polymorphisms and protein structure changes with growth traits and blood metabolites in two Iranian sheep breeds, J. Genet., 93, 831–835, 2014.
Savage, J. J., Yaden, B. C., Kiratipranon, P., and Rhodes, S. J.: Transcriptional control during mammalian anterior pituitary development, Gene, 319, 1–19, https://doi.org/10.1016/S0378-1119(03)00804-7, 2003.
Selvaggi, M. and Dario, C.: Analysis of two Pit-1 gene polymorphisms: Single nucleotide polymorphisms (SNPs) distribution patterns in Podolica cattle breed, Afr. J. Biotechnol., 10, 11360–11364, https://doi.org/10.5897/AJB10.2408, 2011.
Seong, J., Oh, J. D., Cheong, I. C., Lee, K. W., Lee, H. K., Suh, D. S., Jeon, G. J., Park, K. D., and Kong, H. S.: Association between polymorphisms of Myf5 and POU1F1 genes with growth and carcass traits in Hanwoo (Korean cattle), Genes Genom., 33, 425–430, https://doi.org/10.1007/s13258-011-0006-4, 2011.
Sharma, S., Tiwari M., Sharma, D., Singh, S. P., Sharma, A., and Pandey, V.: The study of PIT1/POU1F1 gene polymorphism in Indian Barbari goats using PCR-RFLP methods, Indian Vet. J., 90, 15–16, 2013.
Sun, L. P.: Study on sheep POU1F1 gene polymorphism and body weight traits in Liangshan semi fine wool sheep, Master thesis, Sichuan Agricultural University, Chengdu, 2007.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S.: MEGA6, Molecular Evolutionary Genetics Analysis Version 6.0, available at: http://www.megasoftware.net/ (last access: 10 May 2016), 2013.
Tuggle, C. and Trenkle, A.: Control of growth hormone synthesis, Domest. Anim. Endocrin., 13, 1–33, https://doi.org/10.1016/0739-7240(95)00059-3, 1996.
Wang, J., Li, G., Elzo, M. A., Yan, L., Chen, S., Jia, X., and Lai, S.: A novel single nucleotide polymorphism of the POU1F1 gene associated with meat quality traits in rabbits, Ann. Anim. Sci., 15, 611–620, https://doi.org/10.1515/aoas-2015-0015, 2015.
Woollard, J., Tuggle, C. K., and Ponce de León, F. A.: Localization of POU1F1 to bovine, ovine, and caprine 1q21–22, J. Anim. Sci., 78, 242–243, https://doi.org/10.2527/2000.781242x, 2000.
Yeh, F., Yang, R. C., and Boyle, T.: Popgene (v. 1.32), Microsoft Windows-Based Freeware For Population Genetic Analaysis, available at: http://www.ualberta.ca/~fyeh/PoP32.exe (last access: 15 May 2016) , 2000.
Zhang, C., Liu, B., Chen, H., Lan, X., Lei, C., Zhang, Z., and Zhang, R.: Associations of a HinfI PCR-RFLP of POU1F1 gene with growth traits in Qinchuan cattle, Anim. Biotechnol., 20, 71–74, https://doi.org/10.1080/10495390802640462, 2009.
Zhang, C. F., Chen, H., Zhang, Z. Y., Zhang, L. Z., Yang, D. Y., Qu, Y. J., Hua, L. S., Zhang, B., and Hu, S. R.: A 5′ UTR SNP of GHRHR locus is associated with body weight and average daily gain in Chinese cattle, Mol. Biol. Rep., 39, 10469–10473, https://doi.org/10.1007/s11033-012-1927-y, 2012.
Zhou, F., Yang, Q., Lei, C., Chen, H., and Lan, X.: Relationship between genetic variants of POU1F1, PROP1, IGFBP3 genes and milk performance in Guanzhong dairy goats, Small. Ruminant Res., 140, 40–45, https://doi.org/10.1016/j.smallrumres.2016.05.015, 2016.