the Creative Commons Attribution 3.0 License.
the Creative Commons Attribution 3.0 License.
Identification of a novel 24 bp insertion–deletion (indel) of the androgen receptor gene and its association with growth traits in four indigenous cattle breeds
Haidong Zhao
Mingli Wu
Shuhui Wang
Xiaohui Yu
Ze Li
Ruihua Dang
Xiuzhu Sun
During the past decades, insertions and deletions (indels) have become increasingly popular in animal breeding for understanding the relationship between genotypes and phenotypes. The androgen receptor (AR) plays the vital role of a bridge on the function of the androgen and has sexual size dimorphism. For this reason, the objective of this study was to explore the novel indel variants within the cattle AR gene and to detect their effects on growth traits in four breeds of Chinese yellow cattle. Herein, we first confirmed a novel 24 bp indel (AC_000187.1g.4187270-4187293delAATTTATTGGGAGATTATTGAATT) within the intron of the cattle AR gene. This is consistent with the results predicted from the NCBI SNP database. The distribution of the indel genotypes of four Chinese yellow cattle were significantly different from each other (P < 0.01). After significant correlation analysis, many remarkable phenotypic differences among the three genotypes were found (P < 0.05). In conclusion, a novel 24 bp indel within the AR gene significantly affected growth traits, suggesting that this indel may be a useful DNA marker for the elimination or selection of excellent individuals for cattle breeding.
- Article
(1231 KB) - Full-text XML
- BibTeX
- EndNote
Chinese yellow cattle are transitioning from their previous role as draft cows to beef cattle (Li et al., 2014; Liang et al., 2016). The muscling of their forequarters, which is the key characteristic of draft cows, has been disappearing, and they have taken on many traits characteristic of beef cattle. The important breeds, Qinchuan (QC) cattle, Nanyang (NY) cattle, Luxi (LC) cattle, and Jiaxian red (JX) cattle, have developed a few differences in genotype and phenotype (Liu et al., 2016; Han et al., 2017). It is worth mentioning that QC cattle have developed into a new beef line of QC cattle after nearly 30 years of systematic breeding (Liang et al., 2014; Liu et al., 2015).
Animal growth and development is a complicated process, and the molecular mechanisms of traditional breeding are poorly understood and merit further study. During the past few decades, insertion and deletions (indels) have become increasingly popular in the study of animal breeding for understanding the relationship between genotypes and phenotypes. They are an important form of marker-assisted selection (MAS; Xu et al., 2016; Yang et al., 2016; Zhou et al., 2016).
Androgens are key factors in differences in the growth traits of cows, bulls, and bullocks. They perform a great number of functions by combining with the androgen receptor (AR; O'Reilly et al., 2014). As a member of the nuclear receptor superfamily, the AR could regulate downstream target genes and thereby alter various cell functions. The AR plays the vital role of a bridge for the function of androgens (Myung et al., 2017; Ryan et al., 2017). The AR is a transcription factor that is activated upon binding to testosterone (T), and it is implicated in regulation of the expression of development-related and reproduction-related genes (Grigorova et al., 2017). Some studies have indicated that the AR mediated the expression of androgen-associated somatic traits, such as muscle mass and strength (Ryan et al., 2017).
Table 1PCR primer sequences of the cattle AR gene.
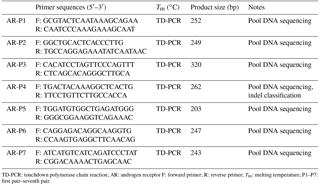
TD-PCR: touchdown polymerase chain reaction; AR: androgen receptor F: forward primer; R: reverse primer; Tm: melting temperature; P1–P7: first pair–seventh pair.
The objective of this study was to explore the novel indel variants within the cattle AR gene and to evaluate their effects on growth traits in four breeds of Chinese yellow cattle, which would not only extend the spectrum of genetic variations in the cattle AR gene but also contribute to implementing MAS in genetics and breeding in Chinese yellow cattle.
All experiments performed in this study were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU). Furthermore, the care and use of animals was fully compliant with local animal welfare laws, guidelines, and policies.
2.1 DNA samples and data collection
A total of 614 Chinese yellow cattle (2–5 years old) from four breeds were measured: Luxi cattle (LX, n= 113, Shandong Province), Qinchuan cattle (QC, n= 227, Shaanxi Province), Nanyang cattle (NY, n= 134, Henan Province), and Jiaxian red cattle (JX, n= 140, Henan Province). Growth traits for all healthy and unrelated individuals, specifically body weight (BW), body height (BH), body length (BL), chest circumference (ChC), chest depth (ChD), chest width (ChW), hucklebone width (HuW), hip width (HW), and cannon circumference (CaC), were measured by the same person using the same standards; consequently, body length index (BLI), chest circumference index (ChCI), chest width index (ChWI), cannon circumference index (CaCI), hucklebone width index (HuWI), and trunk index (TI) were also calculated on the basis of related reported descriptions (Lan et al., 2007; Jin et al., 2016).
2.2 DNA isolation and genomic DNA pool construction
DNA samples were extracted from the leukocytes of the blood by using the phenol–chloroform method (S. H. Zhang et al., 2015; X. Y. Zhang et al., 2015). The quality of DNA samples was assayed by using a NanoDrop 1000 (Thermo Scientific, Waltham, MA, US), and all the data of OD268∕280 remained within a range of 1.8 to 2.0. Every DNA sample was homogenized, and its concentration was diluted to 50 ng µL−1. Considering workloads, every 25 samples were used to construct a genomic DNA pool for polymerase chain reaction (PCR) and sequencing to find a potential indel locus in the cattle AR gene (Chen et al., 2016).
2.3 Primer design and PCR amplification
Based on the SNP database from NCBI (https://www.ncbi.nlm.nih.gov/snp), the loci of seven possible indels were found on the cattle AR intron. The seven pairs of primers were designed by Primer Premier software 5.0 (Premier Biosoft International, USA) based on the cattle AR gene sequence (GenBank NC_ 019460.1; Table 1). The PCR amplification system adopts a 20 µL volume system: 10 µL 2 × PCR mix, 0.5 µM of forward and reverse primers, 50 ng genomic DNA, and 8 µL ddH2O. The protocol for PCR was touchdown PCR, with steps listed as follows: denatured at 95 ∘C for 5 min, followed by 18 cycles of 95 ∘C for 30 s, 68 ∘C declining to 50 ∘C, decreasing one degree per cycle for 30 s and 72 ∘C for 20 s, transition from 68 ∘C to 53 ∘C for 30 s over the course of 20 cycles, followed by final extension at 72 ∘C for 10 min. The products were detected by electrophoresis of 2.5 % agarose gel stained with GelRed (Solarbio Life Science, China), and the products were sequenced only when a given pair of primers had different genotypes (S. H. Zhang et al., 2015; Yang et al., 2016).
2.4 Statistical analyses
The sequence was contrasted and analyzed with BioEdit Software (UK) using the website (http://www.msrcall.com/) to calculate and analyze the genetic data for Hardy–Weinberg equilibrium (HWE), homozygosity (Ho), heterozygosity (He), effective allele numbers (Ne), and polymorphism information content (PIC; Chen et al., 2016). The chi-square test between varieties and ANOVA in varieties was conducted using SPSS software (version 18.0; International Business Machines, US) The results were subjected to statistical testing (P < 0.05; Pan et al., 2013; Zhang et al., 2016).
Through the pooled DNA sequencing and indel classification, only one polymorphism was found in the cattle AR gene (Table 1). After 50 min of agarose gel (2 %) electrophoresis, three genotypes showed different bands (Fig. 1). The II (insertion–insertion) type showed one band (262 bp), the DD (deletion–deletion) type displayed one band (238 bp), and the ID (insertion–deletion) type showed two bands (262 bp, 238 bp). Based on the sequencing results (Fig. 2), genotype II showed two replica and the DD genotypes only had one replica; the del portion is AATTTATTGGGAGATTATTGAATT, the same as the information in the NCBI database (AC_000187.1g.4187270:4187293).
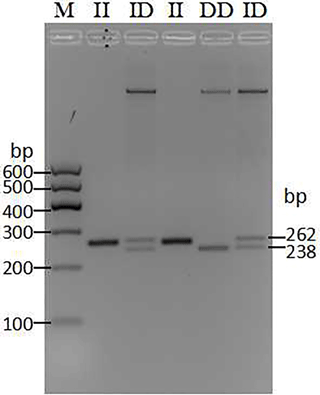
Figure 1The agarose gel electrophoresis patterns of the 24 bp indel within the cattle AR gene. PCR products showed two genotypes at this locus where the insertion–insertion type (II genotype) consisted of 262 bp, deletion–deletion types (DD genotype) consisted of 238 bp, and the heterozygote type (ID genotype) showed 262 and 238 bp, which were detected by 2.5 % agarose gel electrophoresis. M: maker I (600, 500, 400, 300, 200, and 100 bp).
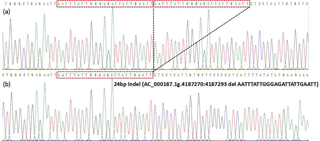
Figure 2Sequencing maps for the 24 bp indel in the cattle AR gene. Panel (a): homozygotic insertion type (II); panel (b): homozygotic deletion type (DD). The sequence with the red border is the 24 bp deletion.
Table 2Genotypes, alleles, He, Ne, and PIC for the novel indel of the cattle AR gene.
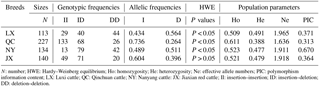
N: number; HWE: Hardy–Weinberg equilibrium; Ho: homozygosity; He: heterozygosity; Ne: effective allele numbers; PIC: polymorphism information content; LX: Luxi cattle; QC: Qinchuan cattle; NY: Nanyang cattle: JX: Jiaxian red cattle; II: insertion–insertion; ID: insertion–deletion; DD: deletion–deletion.
Table 3The χ2 test of different breeds on a novel indel of the cattle AR gene.
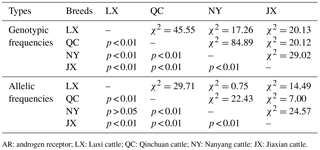
AR: androgen receptor; LX: Luxi cattle; QC: Qinchuan cattle; NY: Nanyang cattle: JX: Jiaxian cattle.
As shown in Table 2, the genotype frequencies, allelic frequencies, and population parameters of four cattle breeds (LX, QC, NY, and JX) were calculated. For the current locus, only JX showed Hardy–Weinberg equilibrium (HWE; P > 0.05). The effective allele numbers (Ne) of LX, JX, and NY were greater than 1.900. The NY cattle were highly polymorphic, and the other breeds had intermediate polymorphism information contents. The four breeds had a different superior genotype; ID was the greater genotype in NY and JX cattle, while DD was the dominant genotype in QC and LX cattle. With the exception of the allelic frequency between LX and NY, the frequency and allelic frequency distribution of all genotypes differed significantly among all four breeds on the χ2 test (P < 0.01; Table 3).
Table 4Relationship between the novel 24 bp indel of the cattle AR gene and growth traits in four cattle breeds.
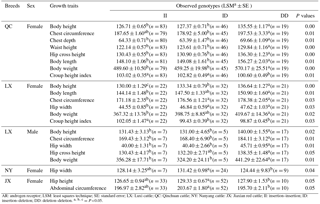
AR: androgen receptor; LSM: least squares technique; SE: standard error; LX: Luxi cattle; QC: Qinchuan cattle; NY: Nanyang cattle: JX: Jiaxian red cattle; II: insertion–insertion; ID: insertion–deletion; DD: deletion–deletion. =P<0.05.
The associations between the 24 bp indel and the cattle growth traits were investigated. Significant differences were found in the body height, chest circumference, chest depth, waist height, hip cross height, body length, body weight, and croup height index of female QC cattle among different genotypes (P < 0.05). Significant differences were observed in body height, body length, chest circumference, hip width, body weight, and croup height index of female LX cattle across three genotypes (P < 0.05). Significant differences were also found in body height, chest circumference, hip width, hip cross height, and body weight of male LX cattle among three genotypes (P < 0.05). Additionally, there is significant difference in the hip width (P=0.04) of female NY cattle among different genotypes. Moreover, hip height (P=0.05) and abdominal circumference (P=0.05) differed significantly among three genotypes in female JX red cattle (Table 4). No significant relationship was observed among growth traits.
The process of animal growth and development is regulated by an extremely complicated network, which contains numerous signaling pathways and genes, including the AR. Recently, many studies have confirmed that the AR is a key hub in the process of animal growth and development, not just in males but also in females (Wu et al., 2017). It is a master regulator that plays a vital role in the regulation of growth and the protection of endangered breeds. Current studies pay more attention to the function of lipometabolism and cancer, placing almost no focus on animal growth and development (Cameron et al., 2016; Ponce-González et al., 2016; Hu et al., 2017; Recouvreux et al., 2017). Herein, we first confirmed the existence of a novel 24 bp indel (AC_000187.1g.4187270-4187293del AATTTATTGGGAGATTATTGAATT) within the intron of cattle AR genes in four breeds of indigenous cattle. This is consistent with that predicted from the NCBI SNP database. Androgens target many organs, including those of the female reproductive pathways (ovaries), male reproductive pathways, bone, the cardiovascular system, the immune system, skin, the kidney, the lungs, fat tissue, and the brain (Usui et al., 2014). Androgens combine with androgen receptors, causing animals to accumulate fat and indirectly regulating growth (O'Reilly et al., 2014). Most importantly, androgens control sexual dimorphism (Pollock et al., 2017).
As shown in the analysis of population genetics data, the number of effective alleles was close to 2 in the LC, NY, and JX cattle. The results showed that this locus had a uniform distribution in the three cattle breeds. According to the standards of polymorphism information content, these four Chinese yellow cattle breeds had intermediate polymorphism information. That is to say, the genetic variation in this site is abundant, which could have a pronounced effect on selective breeding (Li et al., 2016; Ahmad et al., 2017). The genotypic distribution of QC cattle, LX cattle, and JX cattle was not in Hardy–Weinberg equilibrium (HWE) (P > 0.05). This was because these cattle live in a relatively stable environment and had reached a dynamic balance after long-term atresia breeding. However, the other three breeds of cattle were not at HWE (P > 0.05). This could be because of excessive human intervention in the management of livestock farms. The indel genotype distributions of four Chinese yellow cattle breeds were significantly different from each other (P < 0.01), which illustrated the transformation of Chinese cattle from draft cows to beef cattle (François et al., 2017; Mouresan et al., 2017).
After significant correlation analysis, many remarkable phenotypic differences were found. The locus was located in introns, and there is no denying that this could affect animal growth in some way. First, the distance from 5′UTR to exon 1 was more than 100 000 bp, covering a functional area of an unknown gene according to the powerful effect on growth traits (Smith et al., 1977; Lèbre and Gascuel, 2017; Li and Lan, 2015). Second, some studies have confirmed that introns can also affect the expression of genes (Parenteau et al., 2008, 2011; Chorev and Carmel, 2013; Ramke et al., 2017). Third, this locus could affect the expression of the AR gene by regulating alternative splicing (Yang et al., 2017). As this research progresses, it could provide evidence that the combined effects of all of the genomic differences regulate the complex process of animal growth and development. As shown in Table 4, the optimal genotypes were not the same for each breed. The DD genotype was the best genotype in LC and QC cattle, but the ID genotype was the best in NY and JX cattle. This could be due to underpopulation.
In conclusion, we performed a comprehensive study of molecular genetic markers. Ideally, the use of indels will allow us to uncover every gene associated with growth. It could then be combined with the traditional methods of single-strand conformation polymorphism (SSCP), restriction fragment length polymorphism (RFLP), and amplification-created restriction site (ACRS), as well as new methods such as whole-genome resequencing (WGR) and genome-wide association study (GWAS; Tian et al., 2008; Niu et al., 2017; Hibicke et al., 2017; Y. Zhang et al., 2015; Choi et al., 2015; Jiang et al., 2016). Comparatively analyzing the value of molecular markers should be an important choice. Briefly, a novel 24 bp indel within the AR gene significantly affected growth traits, suggesting that this indel may be a potentially useful DNA marker for eliminating or selecting excellent individuals for MAS breeding in cattle.
Our results confirmed the existence of a 24 bp indel within the introns of AR genes in four breeds of Chinese yellow cattle, and we verified their association with growth traits.
Data are available upon request.
XS and RD designed the experiments; HZ, MW, SW, XY, and ZL collected DNA samples; HZ and MW carried out experiments; HZ and MW analyzed the experimental data; and XS and HZ wrote the paper.
The authors declare that they have no conflict of interest.
This work was supported by the Collaborative Innovation Major Projects of
Research and Production Practical Application for Yangling Demonstration
Zone (K3360216067) and the Fundamental Research Funds for the Central
Universities (Z109021505). We would like to thank the staff of the National
Beef Cattle Improvement Center (Shaanxi), the original breeding farm of Jiaxian
red cattle (Henan), the original breeding farm of Nanyang cattle (Henan), and
the reservation farm of Luxi cattle (Shandong) for providing us with samples.
Edited by: Steffen Maak
Reviewed by: two anonymous referees
Ahmad, H. I., Liu, G., Jiang, X., Liu, C., Xu, F. Z., Chong, Y., Ijaz, N., and Huang, H. R.: Adaptive selection at agouti gene inferred breed specific selection signature within the indigenous goat populations, Asian-Australas. J. Anim. Sci., https://doi.org/10.5713/ajas.16.0994, 2017.
Chen, R., Yu, S., Ren, F., Lv, X. Y., and Pan, C. Y.: Detection of one large insertion/deletion (indel) and two novel SNPs within the SPEF2 gene and their associations with male piglet reproduction traits, Arch. Anim. Breed., 59, 275–283, https://doi.org/10.5194/aab-59-275-2016, 2016.
Choi, J. W., Choi, B. H., Lee, S. H., Lee, S. S., Kim, H. C., Yu, D., Chung, W. H., Lee, K. T., Chai, H. H., Cho, Y. M., and Lim, D.: Whole-Genome resequencing analysis of hanwoo and yanbian cattle to identify genome-wide SNPs and signatures of selection, Mol Cells, 38, 466–473, 2015.
Chorev, M. and Carmel, L.: Computational identification of functional introns: high positional conservation of introns that harbor RNA genes, Nucleic Acids Res., 41, 5604–5613, 2013.
Cameron, J. L., Jain, R., Rais, M., White, A. E., Beer, T. M., Kievit, P., Winters-Stone, K., Messaoudi, I., and Varlamov, O.: Perpetuating effects of androgen deficiency on insulin resistance, Int. J. Obes. (Lond)., 40, 1856–1863, 2016.
François, L., Wijnrocx, K., Colinet, F. G., Gengler, N., Hulsegge, B., Windig, J. J., Buys, N., and Janssens, S.: Genomics of a revived breed: Case study of the belgian campine cattle, PLoS One, 20, e0175916, https://doi.org/10.1371/journal.pone.0175916, 2017.
Grigorova, M., Punab, M., Kahre, T., Ivandi, M., Tõnisson, N., Poolamets, O., Vihljajev, V., Žilaitienė, B., Erenpreiss, J., Matulevičius, V., and Laan, M.: The number of CAG and GGN triplet repeats in the androgen receptor gene exert combinatorial effect on hormonal and sperm parameters in young men, Andrology, 5, 495–504, 2017.
Han, Y. J., Chen, Y., Liu, Y., and Liu, X. L.: Sequence variants of the LCORL gene and its association with growth and carcass traits in Qinchuan cattle in China, J. Genet., 96, 9–17, 2017.
Hu, X. Q., Chen, W. L., Ma, H. G., and Jiang, K.: Androgen receptor expression identifies patient with favorable outcome in operable triple negative breast cancer, Oncotarget, 8, 56364–56374, https://doi.org/10.18632/oncotarget.16913, 2017.
Hibicke, M., Graham, M. A., and Hayslett, R. L.: Adolescent chronic restraint stress (aCRS) elicits robust depressive-like behavior in freely cycling, adult female rats without increasing anxiety-like behaviors, Exp. Clin. Psychopharmacol., 25, 74–83, 2017.
Jin, Y., Cai, H., Liu, J., Lin, F., Qi, X., Bai, Y., Lei, C., Chen, H., and Lan, X.: The 10 bp duplication insertion/deletion in the promoter region within paired box 7 gene is associated with growth traits in cattle, Arch. Anim. Breed., 59, 469–476, https://doi.org/10.5194/aab-59-469-2016, 2016.
Jiang, J., Gao, Y., Hou, Y., Li, W., Zhang, S., Zhang, Q., and Sun, D.: Whole-genome resequencing of holstein bulls for indel discovery and identification of genes associated with milk composition traits in dairy cattle, PLoS One, 28, e0168946, https://doi.org/10.1371/journal.pone.0168946, 2016.
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., Lei, C. Z., Zhang, A. L., and Zhang, L: An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits, Small Ruminant Res., 73, 8–12, 2007.
Lèbre, S. and Gascuel, O.: The combinatorics of overlapping genes, J. Theor. Biol., 415, 90–101, 2017.
Liang, W., Zhang, H. L., Liu, Y., Lu, B. C., Liu, X., Li, Q., and Cao, Y.: Investigation of the association of two candidate genes (H-FABP and PSMC1) with growth and carcass traits in Qinchuan beef cattle from China, Genet. Mol. Res., 13, 1876–1884, 2014.
Liang, R. R., Zhu, H., Mao, Y. W., Zhang, Y. M., Zhu, L. X., Cornforth, D., Wang, R. H., Meng, X. Y., and Luo, X.: Tenderness and sensory attributes of the longissimus lumborum muscles with different quality grades from Chinese fattened yellow crossbred steers, Meat Sci., 112, 52–57, 2016.
Li, L., Zhu, Y., Wang, X., He, Y., and Cao, B.: Effects of different dietary energy and protein levels and sex on growth performance, carcass characteristics and meat quality of F1 Angus× Chinese Xiangxi yellow cattle, J. Anim. Sci. Biotechnol., 5, 21–32, 2014.
Li, W. and Lan, P.: Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots, BMC Res Notes, 8, 555–570, https://doi.org/10.1186/s13104-015-1524-y, 2015.
Li, Z., Chen, J., Wang, Z., Pan, Y., Wang, Q., Xu, N., and Wang, Z.: Detection of selection signatures of population-specific genomic regions selected during domestication process in Jinhua pigs, Anim. Genet., 47, 672–681, 2016.
Liu, M., Liu, M., Li, B., Zhou, Y., Huang, Y. Z., Lan, X. Y., Qu, W. D., Qi, X. S., Bai, Y. Y., and Chen, H.: Polymorphisms of FLII implicate gene expressions and growth traits in Chinese cattle, Mol. Cell. Probes., 30, 266–272, 2016.
Liu, Y., Duan, X., Chen, S., He, H., and Liu, X.: NCAPG is differentially expressed during longissimus muscle development and is associated with growth traits in Chinese Qinchuan beef cattle, Genet. Mol. Biol., 38, 450–456, 2015.
Myung, J. K., Wang, G., Chiu, H. H., Wang, J., Mawji, N. R., and Sadar, M. D.: Inhibition of androgen receptor by decoy molecules delays progression to castration-recurrent prostate cancer, PLoS One, 12, e0174134, https://doi.org/10.1371/journal.pone.0174134, 2017.
Mouresan, E. F., Altarriba, J., Moreno, C., Munilla, S., González-Rodríguez, A., and Varona, L.: Performance of genomic selection under a single-step approach in autochthonous Spanish beef cattle populations, J. Anim. Breed Genet., 134, 289–299, https://doi.org/10.1111/jbg.12253, 2017.
Niu, X., Tyasi, T. L., Qin, N., Liu, D., Zhu, H., Chen, X., Zhang, F., Yuan, S., and Xu, R.: Sequence variations in estrogen receptor 1 and 2 genes and their association with egg production traits in Chinese dagu chickens, J. Vet. Med. Sci., 79, 927–934, https://doi.org/10.1292/jvms.17-0014, 2017.
O'Reilly, M. W., House, P. J., and Tomlinson, J. W.: Understanding androgen action in adipose tissue, J. Steroid Biochem. Mol. Biol., 143, 277–284, 2014.
Pan, C. Y., Wu, C. Y., Jia, W. C., Xu, Y., Hu, S. R., Lei, C. Z., Lan, X. Y., and Chen, H.: A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle, Gene, 531, 398–402, 2013.
Parenteau, J., Durand, M., Véronneau, S., Lacombe, A. A., Morin, G., Guérin, V., Cecez, B., Gervais-Bird, J., Koh, C. S., Brunelle, D., Wellinger, R. J., Chabot, B., and Abou Elela, S.: Deletion of many yeast introns reveals a minority of genes that require splicing for function, Mol. Biol. Cell., 19, 1932–1941, 2008.
Parenteau, J., Durand, M., Morin, G., Gagnon, J., Lucier, J. F., Wellinger, R. J., Chabot, B., and Elela, S. A.: Introns within ribosomal protein genes regulate the production and function of yeast ribosomes, Cell, 147, 320–331, 2011.
Pollock, N. B., Feigin, S., Drazenovic, M., and John-Alder, H. B.: Sex hormones and the development of sexual size dimorphism: 5α-dihydrotestosterone inhibits growth in a female-larger lizard (Sceloporus undulatus), J. Exp. Biol., 220, 4068–4077, 2017.
Ponce-González, J. G., Rodríguez-Garcia, L., Losa-Reyna, J., Guadalupe-Grau, A., Rodriguez-Gonzalez, F. G., Díaz-Chico, B. N., Dorado, C., Serrano-Sanchez, J. A., and Calbet, J. A.: Androgen receptor gene polymorphism influence fat accumulation: A longitudinal study from adolescence to adult age, Scand. J. Med. Sci. Sports, 26, 1313–1320, 2016.
Ramke, M., Lee, J. Y., Dyer, D. W., Seto, D., Rajaiya, J., and Chodosh, J.: The 5'UTR in human adenoviruses: leader diversity in late gene expression, Sci. Rep., 7, 618, https://doi.org/10.1038/s41598-017-00747-y, 2017.
Recouvreux, M. V., Wu, J. B., Gao, A. C., Zonis, S., Chesnokova, V., Bhowmick, N., Chung, L. W., and Melmed, S.: Androgen Receptor Regulation of Local Growth Hormone in Prostate Cancer Cells, Endocrinology, 158, 2255–2268, 2017.
Ryan, C. P., Georgiev, A. V., McDade, T. W., Gettler, L. T., Eisenberg, D. T., Rzhetskaya, M., Agustin, S. S., Hayes, M. G., and Kuzawa, C. W.: Androgen receptor polyglutamine repeat length (AR-CAGn) modulates the effect of testosterone on androgen-associated somatic traits in Filipino young adult men, Am. J. Phys. Anthropol., 163, 317–327, https://doi.org/10.1002/ajpa.23208, 2017.
Smith, M., Brown, N. L., Air, G. M., Barrell, B. G., Coulson, A. R., Hutchison, C. A., and Sanger, F.: DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174, Nature, 265, 702–705, 1977.
Tian, D. C., Wang, Q., Zhang, P. F., Araki, H., Yang, S. H., Kreitman, M., Nagylaki, T., Hudson, R., Bergelson, J., and Chen, J. Q.: Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes, Nature, 455, 105–109, 2008.
Usui, T., Kajita, K., Kajita, T., Mori, I., Hanamoto, T., Ikeda, T., Okada, H., Taguchi, K., Kitada, Y., Morita, H., Sasaki, T., Kitamura, T., Sato, T., Kojima, I., and Ishizuka, T.: Elevated mitochondrial biogenesis in skeletal muscle is associated with testosterone-induced body weight loss in male mice, FEBS Lett., 588, 1935–1941, 2014.
Wu, W. F., Maneix, L., Insunza, J., Nalvarte, I., Antonson, P., Kere, J., Yu, N. Y., Tohonen, V., Katayama, S., Einarsdottir, E., Krjutskov, K., Dai, Y. B., Huang, B., Su, W., Warner, M., and Gustafsson J. A.: Estrogen receptor β, a regulator of androgen receptor signaling in the mouse ventral prostate, P. Natl. Acad. Sci. USA, 114, E3816–E3822, https://doi.org/10.1073/pnas.1702211114, 2017.
Xu, L., Hou, Y., Bickhart, D. M., Zhou, Y., Hay el, H. A., Song, J., Sonstegard, T. S., Van Tassell, C. P., and Liu, G. E.: Population-genetic properties of differentiated copy number variations in cattle, Sci Rep., 6, 23161, https://doi.org/10.1038/srep23161, 2016.
Yang, Q., Zhang, S. H., Liu, L., Cao, X., Lei, C. C., Qi, X., Lin, F., Qu, W., Qi, X., Liu, J., Wang, R., Chen, H., and Lan, X. Y.: Application of mathematical expectation (ME) strategy for detecting low frequency mutations: An example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene, Prion, 10, 409–419, 2016.
Yang, D. Y., Tian, J. H., Li, X., Zhang, X., Yu, J. J., Li, S. Y., Huang, Q., Ma, R., Wang, J., and Jiang, M.: Association between the lenth of CAG repeats in exon 1 of the androgen receptor gene and metabolic syndrome in Chinese men, Journal of Xi'an jiaotong University (medical science), 38, 431–436, 2017 (in Chinese).
Zhang, M., Pan, C., Lin, Q., Hu, S., Dang, R., Lei, C., Chen, H., and Lan, X.: Exploration of the exonic variations of the iPSC-related Nanog gene and their effects on phenotypic traits in cattle, Arch. Anim. Breed., 59, 351–361, https://doi.org/10.5194/aab-59-351-2016, 2016.
Zhang, S. H., Sun, K. A., Bian, Y. N., Zhao, Q., Wang, Z., Ji, C. N., and Li, C. T.: Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China, Sci. Rep., 5, 1833–1836, 2015.
Zhang, X. Y., Wu, X. F., Jia, W. C., Pan, C. Y., Li, X. C., Lei, C. Z., Chen, H., and Lan, X. Y.: Novel Nucleotide Variations, Haplotypes Structure and Associations with Growth Related Traits of Goat AT Motif-Binding Factor (ATBF1) Gene, J. Anim. Sci., 28, 1394–1406, 2015.
Zhang, Y., Guo, G., Huang, H., Lu, L., Wang, L., Fang, L., Liu, L., Wang, Y., and Zhang, S.: Screening for JH1 genetic defect carriers in Jersey cattle by a polymerase chain reaction and restriction fragment length polymorphism assay, J. Vet. Diagn. Invest., 27, 596–599, 2015.
Zhou, Y., Utsunomiya, Y. T., Xu, L., Hay el, H. A., Bickhart, D. M., Alexandre, P. A., Rosen, B. D., Schroeder, S. G., Carvalheiro, R., de Rezende Neves, H. H., Sonstegard, T. S., Van Tassell, C. P., Ferraz, J. B., Fukumasu, H., Garcia, J. F., and Liu, G. E.: Genome-wide CNV analysis reveals variants associated with growth traits in Bos indicus, BMC Genomics, 17, 419, https://doi.org/10.1186/s12864-016-2461-4, 2016.





