the Creative Commons Attribution 3.0 License.
the Creative Commons Attribution 3.0 License.
The effect of short/branched chain acyl-coenzyme A dehydrogenase gene on triglyceride synthesis of bovine mammary epithelial cells
Ping Jiang
Xibi Fang
Zhihui Zhao
Xianzhong Yu
Boxing Sun
Haibin Yu
Short/branched chain acyl-CoA dehydrogenase (ACADSB) is a member of the acyl-CoA dehydrogenase family of enzymes that catalyze the dehydrogenation of acyl-CoA derivatives in the metabolism of fatty acids. Our previous transcriptome analysis in dairy cattle showed that ACADSB was differentially expressed and was associated with milk fat metabolism. The aim of this study was to elucidate the background of this differential expression and to evaluate the role of ACADSB as a candidate for fat metabolism in dairy cattle. After analysis of ACADSB mRNA abundance by qRT-PCR and Western blot, overexpression and RNA interference (RNAi) vectors of ACADSB gene were constructed and then transfected into bovine mammary epithelial cells (bMECs) to examine the effects of ACADSB on milk fat synthesis. The results showed that the ACADSB was differentially expressed in mammary tissue of low and high milk fat dairy cattle. Overexpression of ACADSB gene could significantly increase the level of intracellular triglyceride (TG), while ACADSB gene knockdown could significantly reduce the TG synthesis in bMECs. This study suggested that the ACADSB was important in TG synthesis in bMECs, and it could be a candidate gene to regulate the metabolism of milk fat in dairy cattle.
- Article
(1403 KB) - Full-text XML
- BibTeX
- EndNote
ACADSB gene is a short/branched chain acyl-coenzyme a dehydrogenase, which is a member of the ACADS family that is indispensable in fatty acid and amino acid metabolism in mitochondria (Ikeda et al., 1985; Alfardan et al., 2010). ACADS also catalyzes α, β-dehydrogenation of acyl-CoA esters, resulting in transferring the electron to flavoprotein (Rozen et al., 1994). ACADSB is expressed in adipose tissue, lymphatic reticulum, breast tissue, muscle tissue and rumen. Acyl-CoA dehydrogenases with specificity for different chain lengths of fatty acids in the mitochondria carry out the first committed step of β-oxidation reaction (Ensenauer et al., 2005). Studies revealed that ACADSB gene single-nucleotide polymorphisms are associated with hyperlipidemia in humans (Kamide et al., 2007). ACADSB exhibited key differences in substrate specificity between rats and humans. Despite the overall amino acid identity of 85 %, rat ACADSB is more active than human ACADSB with substrates with longer carbon side chains, but the human enzyme can utilize substrates with the longer primary carbon chains. These residues are in the same position in rat and human ACADSB, indicating the other amino acid residues contribute to the substrate specificity of ACADSB (Ca et al., 2007; Luis et al., 2011; Van Calcar et al., 2013). In humans, ACADSB deficiency from autosomal recessive disorder affects isoleucine catabolism (Nguyen et al., 2002) but not valine catabolism, which has no detectable activity with isobutyryl-CoA (van Calcar et al., 2009; Liu et al., 2013). So far, the role of ACADSB in fat metabolism of cattle has not been extensively studied. However, the effects of ACADSB gene on bovine mammary epithelial cells are rarely reported, especially the function of ACADSB gene on milk fat metabolism. Triglyceride (TG), as important content of very-low-density lipoprotein (VLDL) and chylomicrons, plays a very important role in metabolism as the transporters of fat, and TG is a main constituent of body fat in animals. Fatty acids are essential energy substrates and are also precursors for membrane lipids. In cells, fatty acids are stored in TG within cytoplasmic liquid droplets (Das et al., 2015). Therefore, the content of TG in bovine mammary epithelial cells (bMECs) might show a relationship to milk fat synthesis in dairy cattle. Our previous transcriptome analysis in dairy cattle showed that ACADSB was differentially expressed (Table 1), and we aimed to further explore the functions of ACADSB expression in bMECs of Chinese Holstein cows. We designed overexpression vector and RNA interference (RNAi) vector of ACADSB gene to validate its function on synthesis of TG in fatty-acid metabolism pathway.
Table 1Partial data about the differentially expressed genes of transcriptome analysis.
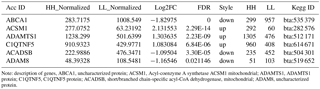
Note: description of genes, ABCA1, uncharacterized protein; ACSM1, Acyl-coenzyme A synthetase ACSM1 mitochondrial; ADAMTS1, ADAMTS1 protein; C1QTNF5, C1QTNF5 protein; ACADSB, short/branched chain-specific acyl-CoA dehydrogenase, mitochondrial; ADAM8, uncharacterized protein.
Animal experiments were performed in strict accordance with the guide for the care and use of laboratory animals by the Jilin University animal care and use committee (permit number: SYXK (Ji) 2012-0010/0011).
2.1 Expression of ACADSB gene in various bovine mammary tissues
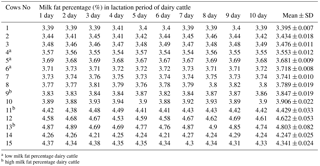
In this study, 3 high milk fat cattle and 3 low milk fat cattle were chosen by a milk ingredient analyzer (Lactoscan SP, Milkatronic Ltd., Nova Zagora, Bulgaria) from 15 healthy dairy cattle, which were in the same environment and the same lactation period. Dairy cattle of different experimental groups were slaughtered at Haoyue slaughterhouse in Changchun, and then the fresh breast tissues were collected from the carcass of dairy cattle with different milk fat percentage and frozen in liquid nitrogen for subsequent experiments. The total RNA was extracted from mammary tissue using TRIzol Reagent (Tiangen, Beijing, China) and reverse transcribed into cDNA using a RT cDNA synthesis kit (TOYOBO, Japan) and following the manufacturer's protocols. ACADSB gene expression in mammary tissues from cattle with different milk fat percentages was analyzed by quantitative real-time fluorescent PCR (qRT-PCR). The qRT-PCR was carried out using SYBR® Premix Ex taqTM (TAKARA). PCR amplification cycles for standard curve were performed as follows: 95 ∘C for 5 min; 40 cycles of 95 ∘C for 30 s and 60 ∘C for 30 s.
2.2 Construction of interference vector pGPU6/GFP/Neo-ACADSB
A 21-mer siRNA oligonucleotide targeting the coding sequence of ACADSB and the complementary oligonucleotide were designed according to the design principles of shRNA. The target sequence on ACADSB gene is GCTGGGTATAAAGGAA TTACC and the sequences of the primer pair were as follows: (5′-CACCGCTGGGTATAAA GGAATTACCTTCAAGAGAGGTAATTCCTTTATACCC AGCTTTTTTG-3′) and (5′-GATCCAAAAAAGCTGGGTATAAAGGAATTA CCTCTCTTGAAGGTAATTC CTTTATACCCAGC-3′). Annealed siRNA oligonucleotides were cloned into pGPU6/GFP/Neo vector (GenePharma Corporation, Shanghai). The 10 µL annealing mixture was as follows: 1 µL annealing buffer, 1 µL sense siRNA template oligonucleotide (100 µM), and 1 µL antisense siRNA template oligonucleotide (100 µM), 7 µL ddH2O.
Negative control plasmid was constructed by inserting annealed scrambled siRNA oligonucleotides into pGPU6/GFP/Neo.
2.3 Construction of overexpression vector pBI-CMV3-ACADSB
The cDNA for the predicted mature coding region of ACADSB in cow was amplified by PCR with sense primer 5′-GGATCCATGGAGCGAGCTACTGTT CGG-3′ containing a BamHI site and antisense primer 5′-ATCGATTCAGTATTCCGC ACTGATGCAC-3′ containing a ClaI site. The 1299 bp PCR products were purified and ligated into pMD18-T simple vector (TAKARA, Beijing, China). The ACADSB sequence that includes cleavage sites of BamHI and ClaI restriction enzyme was ligated into the expression vector of pBI-CMV3. The resultant plasmid, pBI-CMV3-ACADSB, was confirmed by double restriction enzyme digestion with BamHI (NEB, UK) and ClaI (NEB, UK), and further validated with sequencing by Shanghai Sangon Biotech.
2.4 Transfection of plasmids into bovine mammary epithelial cells
The bovine mammary epithelial cells in this study were established by our laboratory (Lu et al., 2012). Cells were cultured in 10 cm culture plates (Falcon, Franklin, Lake, NJ, USA) in DMEM/F12 (HyClone, USA) with 10 % FBS (fetal bovine serum, Invitrogen, USA), 50 µL hydrocortisone (50 µg mL−1, Abcam, UK) and 50 µL insulin (0.5 EU mg−1, Abcam, UK) with 1 % antibiotics (HyClone). Then, bMECs were seeded into six well-culture plates (Falcon, Franklin, Lake, NJ, USA) at a concentration of 0.6 × 106 cells/well and cultured at 37 ∘C in CO2 incubator (Thermo, Marietta, OH, USA). When cells reached at 70 % confluency, cells changed the medium without antibiotics for transfection. Plasmid DNA was transfected into bMECs using FuGENE® HD Transfection Reagent (Promega, USA) according to the manufacturer's instructions. The expression of GFP in transfected cells was observed under a fluorescence microscope (NikonTE2000, Japan) after transfection for 24 h. The transfected cells groups included overexpression group (pBI-CMV3-ACADSB plasmid and pBI-CMV3 control plasmid) and RNAi group (pGPU6/GFP/Neo-ACADSB plasmid and pGPU6/GFP/Neo-shNC control plasmid).
2.5 Analysis of ACADSB expression by quantitative RT-PCR
BMECs transfected with ACADSB RNAi vector and overexpression vector were harvested after transfection for 24 h, and total RNA was extracted using the PREP RNA mini kit (Analytik Jena, Germany). Total RNA was checked with agarose gel electrophoresis, and the concentration was measured using a spectrophotometer. The cDNA was synthesized using a RT cDNA synthesis kit (ToYoBo, Japan) following the manufacturer's protocols. The qRT-PCR was carried out using SYBR® Premix Ex taqTM (TAKARA) and a pair of specific primers ACADSB-F: 5′AGCTACTGTTCGGCTGCTGC 3′, and ACADSB-R: 5′TCCATTGCTTGTCG CTTTGAG 3′. The mRNA expression level was determined by qPCR, and β-actin was served as the reference gene. The relative expression levels of ACADSB mRNA were calculated by 2−ΔΔct method, and the experiment was repeated three times.
2.6 Analysis of ACADSB protein expression by Western blot
The cells were collected after transfection for 48 h and washed two times with PBS (phosphate buffered saline). Cells were then resuspended in RIPA buffer (RIPA splitting buffer, BOSTER, China) with protease inhibitor. Cell lysates were cleared by centrifugation at 12 000 g for 5 min at 4 ∘C, and then the supernatants were collected. The total protein concentration in the samples was determined with the enhanced BCA protein quantitation assay (KeyGEN BioTECH, China) by a spectrophotometer (UNIC2802H, Shanghai, China). For Western blot, equal amounts of various proteins were resolved by SDS-PAGE and then transferred onto PVDF membranes (Bio-Red laboratories Inc, USA) by a trans-blot (SEMI-DRY TRANSFER CELL, BIO-RAD, USA) for 8 min. After blocked with 1 × TBS buffer containing 5 % BSA (bovine serum albumin) for 3 h, the PVDF membrane was washed with TBST solution. Membrane was then incubated with rabbit polyclonal anti-ACADSB antibody (dilution in 1 × TBS, 1:750 Abcam, USA) and β-actin (dilution in 1 × TBS, 1:10 000 Abcam, USA) for 12 h at 4∘. After washing with TBST, membrane was incubated with anti-rabbit secondary antibody (dilution in 1× TBS, 1:2000, BioWorld, USA) for 2 h. The protein bands were visualized with an enhanced chemiluminescent HRP substrate (Thermo, USA). The relative expressions of target protein in different samples were calculated with the gray value analyzer.
2.7 Determination of the triglyceride content in transfected cells
The TG contents were determined using TG kit (Applygen Technologies, Beijing, China) according to the manufacturer's protocols. BME cells were plated at a concentration of 0.6 × 106 cells/well on six-well plates (Falcon, Franklin Lakes, NJ, USA), and the cells were collected after transfection for 48 h with CMV3-ACADSB and pGPU6/GFP/Neo-ACADSB plasmid. Different samples were adjusted to the same concentration, and each sample was repeated three times. Cellular content of TG was adjusted based on the quantity of protein. Total protein concentration was estimated by the BCA protein content detection kit (KeyGen, Biotech, China), and the cellular content of TG was corrected for protein concentration of each microgram. The concentration of TG was determined by a microplate reader (Yong Chuang SM600, Shanghai, China).
2.8 Statistical analysis
Experimental data are shown as mean ± SE. Statistically significant differences are defined as p<0.05. Data from fluorescence quantitative PCR on relative gene expression were analyzed using the comparative Ct method (2−ΔΔCt). Data analysis was performed with SPSS 13.0 software and GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA) using a two-tailed t test (unpaired t test).
3.1 ACADSB expression in various bovine mammary tissues
Cattle short/branched chain acyl-CoA dehydrogenase gene is located on bovine (BTA) chromosome 26 (26:43 134 415–43 184 345). The ACADSB DNA sequence contains 49 930 base pairs, 11 exons and 10 introns (Fig. 2a). ACADSB mRNA sequence contains a 1299 bp coding region (CDS) and a 559 bp 5′, 3′ UTR. The ORF is encoded for a 432-amino-acid protein.
The ACADSB gene expression in mammary tissues from the three high milk fat cattle and three low milk fat cattle was determined by quantitative real-time PCR. The expression of ACADSB gene in mammary tissues of high milk fat cattle is significantly higher than that in the low milk fat cattle (p<0.05, Fig. 1, Table 2).
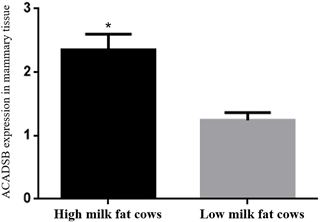
Figure 1The ACADSB gene expression levels in mammary tissue of high milk fat and low milk fat cattle.
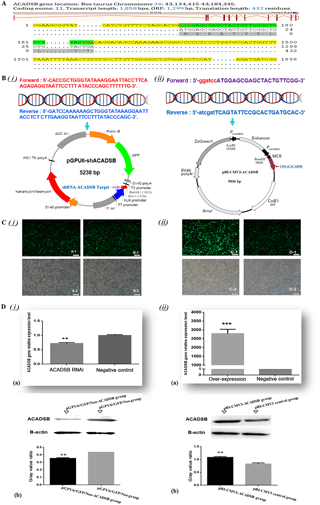
Figure 2Construction of ACADSB gene RNA interference and overexpression vector and transfection into bovine mammary epithelial cells.(A) Bovine ACADSB gene location and structure features. (B) Primer sequence and vector of ACADSB gene. (i) Primer sequence of RNA interference target sequence and interference vector (pGPU6/GFP/Neo-ACADSB). (ii) Primer sequence of ORF and overexpression vector (pBI-CMV3-ACADSB). (C) Plasmid transfection into bovine mammary epithelial cells (A-1) cells transfected with ACADSB RNAi plasmid pGPU6/GFP/Neo-ACADSB, B-1: cells transfected with pGPU6/GFP/Neo control plasmid, C-1: cells transfected with ACADSB overexpression plasmid pBI-CMV3-ACADSB, D-1: cells transfected with pBI-CMV3 control vector. (D) mRNA and protein of ACADSB expression in bovine mammary epithelial cells that transfected with RNAi and overexpression plasmid. (i-a): ACADSB mRNA expression in bMECs that transfected with RNAi plasmid; (i-b): ACADSB protein expression in bMECs that transfected with RNAi plasmid; (ii-a): ACADSB mRNA expression in bMECs that transfected with overexpression plasmid; (ii-b): ACADSB protein expression in bMECs that transfected with overexpression plasmid (the protein expression level of ACADSB using gray value analyzer to calculate gray value ratios (target protein content/β-actin content).
3.2 Transfection of plasmids into bovine mammary epithelial cells
In order to reveal the effect of ACADSB gene on the synthesis of TGs, overexpression vector and RNAi vector of bovine ACADSB gene were successfully constructed (Fig. 2b). Bovine mammary epithelial cells were transiently transfected with plasmids of pGPU6/GFP/Neo-ACADSB and pBI-CMV3-ACADSB. The expression of green fluorescence could be observed from both overexpression vector (pBI-CMV3-ACADSB) transfected group and RNAi vector (pGPU6/GFP/Neo-ACADSB) transfected group after 24 h transfection but not in untransfected group (Fig. 2c).
3.3 Analysis of ACADSB mRNA expression in transfected group by qRT-PCR
To investigate the effect of pBI-CMV3-ACADSB and pGPU6/GFP/Neo-ACADSB vector on cellular ACADSB mRNA level, total RNA was extracted from cells which transfected with various plasmids and analyzed by qRT-PCR. The results showed that ACADSB mRNA expression levels in RNAi transfected cells were significantly decreased compared with those in the negative control vector transfected cells (p<0.05, Fig. 2D(i)–(a)). In the meantime, the ACADSB mRNA expression levels in overexpression transfected cells were significantly increased compared with those in the control vector transfected cells (p<0.01, Fig. 2D(ii)–(a)).
3.4 Expression of ACADSB protein in bMEC transfected with different plasmid
To investigate the effect of pBI-CMV3-ACADSB and pGPU6/GFP/Neo-ACADSB plasmid on cellular ACADSB protein level, total protein was isolated from cells that transfected with various plasmids of overexpression vector and RNAi vector. The Western blot analysis results revealed that ACADSB protein expression in RNAi transfected cells was significantly down-regulated compared to those in the negative control vector transfected cells (p<0.05, Fig. 2D(i)–(b)). Meanwhile, the protein level of ACADSB was significant increased in pBI-CMV3-ACADSB vector transfected group compared with that in the control vector transfected group (p<0.05, Fig. 2D(ii)–(b)). β-actin was used as an internal control. Western blot protein bands were digitized with a gray value analyzer, and ACADSB protein level was expressed as the ratio over β-actin.
3.5 Detection of the triglyceride content in bMEC transfected with different plasmid
To examine the effect of ACADSB gene on intracellular TG synthesis, TG content was detected by TG kit. The result showed that TG content in bMEC transfected with pGPU6/GFP/Neo-ACADSB was significantly lower than that in the control group and pBI-CMV3-ACADSB transfected group (p<0.05, Fig. 3a). Meanwhile, the TG content in bMEC transfected with overexpression vector was significantly higher compared to that in the control vector transfected group (p<0.05, Fig. 3b).
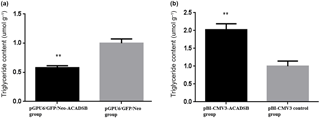
Figure 3Comparison of the TG content of bMECs which transfected with RNAi vector or overexpression vector. (a) The TG content of bMECs which transfected with pGPU6/GFP/Neo-ACADSB vector and pGPU6/GFP/Neo control vector. (b) The TG content of bMECs which transfected with overexpression plasmid pBI-CMV3-ACADSB and pBI-CMV3 control vector.
ACADSB is a member of the acyl-CoA dehydrogenase family, which can catalyze the dehydrogenation of acyl-CoA derivatives in the metabolism of fatty acid (Willard et al., 1996; Arden et al., 1995). At least nine members of this family have been described (Ensenauer et al., 2005; Ikeda et al., 1985). Substrate specificity is the most important characteristic to define the members of the ACADS family. Studies have shown that SCAD, ACADSB and IBD can all utilize butyryl-CoA as a substrate, whereas IVD, ACADSB and IBD are most active with short/branched chain acyl-CoAs as a substrate (Zhang et al., 2002; He et al., 2003). ACADSB encodes for a mitochondrial precursor protein with a molecular weight of 47.1 kDa, which has the active activity toward the short/branched chained acyl-CoA derivative (Rozen et al., 1994; Alfardan et al., 2010). ACADSB gene is located on chromosome 10 in human at 10q26.13 (Andresen et al., 2000). ACADSB is differentially expressed among different tissues (Gibson et al., 2000). Studies in human dehydrogenase deficiency have indicated the involvement of ACADSB gene in the degradation of valine, leucine and isoleucine and the metabolism of fatty-acid metabolism (Chaudhari et al., 2016; Cheng et al., 2016; Wang et al., 2015), suggesting that ACADSB gene maybe a candidate gene of fatty-acid metabolism.
Since the pathway of glycerolipid biosynthesis was illuminated in the 1950s, more studies have been gained about the mechanism of lipid biosynthetic and triacylglycerol biosynthesis (Coleman and Lee, 2004). TG has a critical effect on energy store and a repository of essential and nonessential fatty acids and also was the precursor of phospholipid biosynthesis. Fatty acids are packaged in very low-density lipoprotein and chylomicra as TG for distribution to other tissues (Coleman and Lee, 2004). Fatty-acid composition of TGs affects the absorption and the distribution in the organism (Rodríguez et al., 2012). A published study showed that dietary TG may influence lipid metabolism in humans, and it also found enhanced absorption of fatty acid in the sn-2 position of dietary TGs (Hunter, 2001). Lipogenesis resulted in cellular lipid accumulation, via the uptake of lipogenic substrate. Then the endogenous fatty acid was synthesis and mainly stored in form of TGs (Desvergne et al., 2006). TG is also an essential component of milk (Coleman and Lee, 2004), suggesting that ACADSB gene maybe a candidate gene of milk fat metabolism.
In this study, we have examined the milk fat rate in 15 Chinese Holstein cattle with the same lactation period and compared ACADSB gene expression levels in mammary tissue by quantitative RT-PCR between dairy cattle groups of high fat rates and low milk fat percentage. In order to further validate the function of ACADSB gene, we cloned ACADSB gene from bovine tissue by RT-PCR and constructed the silencing vector and overexpression vector of ACADSB, which were then transfected into bMEC. As a result of our experiment, the ACADSB can significantly affect the synthesis of TGs in the fatty-acid metabolic pathway in bMEC.
The pBI-CMV3 vector was selected to construct the overexpression vector, which is bidirectional mammalian expression vector and is designed to constitutively express a protein of interest and the green fluorescent protein ZsGreen1, and it also can be transfected into mammalian cells using any standard transfection methods (Yu et al., 2017). Therefore, it greatly improves the test process and also ensures the well-off completion of the study. In cell transfection, we recommend the following: when the bMEC confluence is above 80 %, the use of FuGENE transfection reagent could improve transfection efficiency.
In conclusion, our previous transcriptome analysis and current study results identified ACADSB as a differentially expressed gene in mammary tissue between low and high milk fat cattle. Our results showed that ACADSB expression level was positively correlated with cellular TGs content, and ACADSB gene was actively involved in fatty-acid metabolism pathway in bMEC and milk fat synthesis of dairy cattle.
Data are available from the corresponding author upon request.
The authors declare that they have no conflict of interest.
This work was supported by the National Major Special Project on New
Varieties Cultivation for Transgenic Organisms (2016ZX08009003006),
the National Natural Science Foundation of China (no. 31672389,
31772562) and the Jilin Scientific and Technological Development
Program (no. 20170519014JH, 20180101275JC).
Edited by: Steffen Maak
Reviewed by: Hong Guo and one anonymous referee
Alfardan, J., Mohsen, A. W., Copeland, S., Ellison, J., Keppen-Davis, L., Rohrbach, M., Powell, B. R., Gillis, J., Matern, D., Kant, J., and Vockley, J.: Characterization of new ACADSB gene sequence mutations and clinical implications in patients with 2-methylbutyrylglycinuria identified by newborn screening, Mol. Genet. Metab., 100, 333–338, https://doi.org/10.1016/j.ymgme.2010.04.014, 2010.
Andresen, B. S., Christensen, E., Corydon, T. J., Bross, P., Pilgaard, B., Wanders, R. J., Ruiter, J. P., Simonsen, H., Winter, V., Knudsen, I., Schroeder, L. D., Gregersen, N., and Skovby, F.: Isolated 2-methylbutyrylglycinuria caused by short/branched-chain acyl-CoA dehydrogenase deficiency: identification of a new enzyme defect, resolution of its molecular basis, and evidence for distinct acyl-CoA dehydrogenases in isoleucine and valine metabolism, Am, J. Hum. Genet., 67, 1095–1103, https://doi.org/10.1086/303105, 2000.
Arden, K. C., Viars, C. S., Fu, K., and Rozen, R.: Localization of short/branched chain acyl-CoA dehydrogenase (ACADSB) to human chromosome 10, Genomics, 25, 743–745, 1995.
Ca, C., Cl, C., Leuzzi, V., Artiola, C., Santagata, S., De Luca, T., Vestri, L., and Antonozzi, I.: A new case of short/branched-chain acyl-CoA dehydrogenase deficiency caused by two novel mutations, J. Inherit. Metab. Dis., 30, 43–43, 2007.
Chaudhari, A., Pathakota, G. B., and Annam, P. K.: Design and construction of shrimp antiviral DNA vaccines expressing long and short hairpins for protection by RNA interference, Methods in molecular biology, 1404, 225–240, https://doi.org/10.1007/978-1-4939-3389-1_16, 2016.
Cheng, A., Zhang, Y., Mei, H., Fang, S., Ji, P., Yang, J., Yu, L., and Guo, W.: Construction of recombinant pEGFP-N1-hPer2 plasmid and its expression in osteosarcoma cells, Oncol. Lett., 11, 2768–2772, https://doi.org/10.3892/ol.2016.4291, 2016.
Coleman, R. A. and Lee, D. P.: Enzymes of triacylglycerol synthesis and their regulation, Prog. Lipid Res., 43, 134–176, 2004.
Das, S. K., Stadelmeyer, E., Schauer, S., Schwarz, A., Strohmaier, H., Claudel, T., Zechner, R., Hoefler, G., and Vesely, P. W.: Micro RNA-124a regulates lipolysis via adipose triglyceride lipase and comparative gene identification 58, Int. J. Mol. Sci., 16, 8555–8568, 2015.
Desvergne, B., Michalik, L., and Wahli, W.: Transcriptional regulation of metabolism, Physiol. Rev., 86, 465–514, 2006.
Ensenauer, R., He, M., Willard, J. M., Goetzman, E. S., Corydon, T. J., Vandahl, B. B., Mohsen, A. W., Isaya, G., and Vockley, J.: Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids, J. Biol. Chem., 280, 32309–32316, https://doi.org/10.1074/jbc.M504460200, 2005.
Gibson, K. M., Burlingame, T. G., Hogema, B., Jakobs, C., Schutgens, R. B., Millington, D., Roe, C. R., Roe, D. S., Sweetman, L., Steiner, R. D., Linck, L., Pohowalla, P., Sacks, M., Kiss, D., Rinaldo, P., and Vockley, J.: 2-Methylbutyryl-coenzyme A dehydrogenase deficiency: a new inborn error of L-isoleucine metabolism, Pediatr. Res., 47, 830–833, 2000.
He, M., Burghardt, T. P., and Vockley, J.: A novel approach to the characterization of substrate specificity in short/branched chain Acyl-CoA dehydrogenase, J. Biol. Chem., 278, 37974–37986, https://doi.org/10.1074/jbc.M306882200, 2003.
Hunter, J. E.: Studies on effects of dietary fatty acids as related to their position on triglycerides, Lipids, 36, 655–668, 2001.
Ikeda, Y., Okamura-Ikeda, K., and Tanaka, K.: Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme, J. Biol. Chem., 260, 1311–1325, 1985.
Kamide, K., Kokubo, Y., Yang, J., Matayoshi, T., Inamoto, N., Takiuchi, S., Horio, T., Miwa, Y., Yoshii, M., Tomoike, H., Tanaka, C., Banno, M., Okuda, T., Kawano, Y., and Miyata, T.: Association of genetic polymorphisms of ACADSB and COMT with human hypertension, J. Hypertens., 25, 103–110, https://doi.org/10.1097/HJH.0b013e3280103a40, 2007.
Liu, X. J., Wu, L., Deng, G. S., Chen, G., Li, N., Chu, X. S., and Li, D.: Comparative studies of Acyl-CoA dehydrogenases for monomethyl branched chain substrates in amino acid metabolism, Bioorg. Chem., 47, 1–8, 2013.
Lu, C., Yang, R., Liu, B., Li, Z., Shen, B., Yan, S., Zhang, Y., Zhang, L., and Zhao, Z.: Establishment of two types of mammary epithelial cell lines from chinese holstein dairy cow, J. Anim. Vet. Adv., 11, 1166–1172, 2012.
Luis, P. B. M., Ruiter, J. P. N., IJlst, L., de Almeida, I. T., Duran, M., Mohsen, A., Vockley, J., Wanders, R. J. A., and Silva, M. F. B.: Role of isovaleryl-CoA dehydrogenase and short branched-chain Acyl-CoA dehydrogenase in the metabolism of valproic acid: implications for the branched-chain amino acid oxidation pathway, Drug. Metab. Dispos., 39, 1155–1160, 2011.
Nguyen, T. V., Andresen, B. S., Corydon, T. J., Ghisla, S., Abd-El Razik, N., Mohsen, A. W., Cederbaum, S. D., Roe, D. S., Roe, C. R., Lench, N. J., and Vockley, J.: Identification of isobutyryl-CoA dehydrogenase and its deficiency in humans, Mol. Genet. Metab., 77, 68–79, 2002.
Rodríguez, A., Esteban, L., Martín, L., Jiménez, M. J., Hita, E., Castillo, B., González, P. A., and Robles, A.: Synthesis of 2-monoacylglycerols and structured triacylglycerols rich in polyunsaturated fatty acids by enzyme catalyzed reactions, Enzyme Microb. Tech., 51, 148–155, 2012.
Rozen, R., Vockley, J., Zhou, L., Milos, R., Willard, J., Fu, K., Vicanek, C., Low-Nang, L., Torban, E., and Fournier, B.: Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family, Genomics, 24, 280–287, https://doi.org/10.1006/geno.1994.1617, 1994.
van Calcar, S. C., Baker, M. W., Jones, S., Kopish, G., Lee, M., Lee, S., Hoffman, G., Vockley, G., Rhead, W. J., Rice, G. M., and Durkin, M. S.: Mutation analysis and incidence of 2-methylbutyryl-Coa dehydrogenase deficiency (short/branched-chain Acyl-CoA dehydrogenase deficiency; Sbcadd) in the Wisconsin Hmong Population, Mol. Genet. Metab., 98, 112–112, 2009.
Van Calcar, S. C., Baker, M. W., Williams, P., Jones, S. A., Xiong, B., Thao, M. C., Lee, S., Yang, M. K., Rice, G. M., Rhead, W., Vockley, J., Hoffman, G., and Durkin, M. S.: Prevalence and mutation analysis of short/branched chain acyl-CoA dehydrogenase deficiency (SBCADD) detected on newborn screening in Wisconsin, Mol. Genet. Metab., 110, 111–115, 2013.
Wang, J., Liang, H., Zhao, Y., Liu, X., Yang, K., and Sui, A.: Construction and identification of an RNA interference lentiviral vector targeting the mouse TNF-alpha gene, Exp, Ther. Med., 10, 2283–2288, https://doi.org/10.3892/etm.2015.2813, 2015.
Willard, J., Vicanek, C., Battaile, K. P., Van Veldhoven, P. P., Fauq, A. H., Rozen, R., and Vockley, J.: Cloning of a cDNA for short/branched chain acyl-Coenzyme A dehydrogenase from rat and characterization of its tissue expression and substrate specificity, Arch. Biochem. Biophys., 331, 127–133, https://doi.org/10.1006/abbi.1996.0290, 1996.
Yu, H., Zhao, Z., Yu, X., Li, J., Lu, C., and Yang, R.: Bovine lipid metabolism related gene GPAM: molecular characterization, function identification, and association analysis with fat deposition traits, Gene, 609, 9–18, https://doi.org/10.1016/j.gene.2017.01.031, 2017.
Zhang, J., Zhang, W., Zou, D., Chen, G., Wan, T., Zhang, M., and Cao, X.: Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family, Biochem. Bioph. Res. Co., 297, 1033–1042, 2002.